Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria
Figures

H-NS formed two different filaments depending on concentration.
(A) The H-NS-silenced E. coli bgl operon, encoding genes for ß-glucoside catabolism, contains an antisense promoter within bglF (PAS) (Peters et al., 2012). The 1.56-kb linear λPR-bgl DNA template contains the λPR promoter followed by a 26-nucleotide C-less cassette (to allow formation of halted A26 ECs) and two high affinity H-NS binding sites (DRE and URE) (Dole et al., 2004). Native PAGE of H-NS filaments on 10 pM or 10 nM λPR-bgl template in 8 mM Mg2+. Graphics depicting bridged and linear H-NS filaments are shown left of the gel and related to H-NS molecular structures in Figure 1—figure supplement 1. (B) Native PAGE of H-NS filaments formed on free DNA or halted A26 complexes (at 10 nM) at 2 or 8 mM Mg2+ and 66 H-NS/kb or 200 H-NS/kb (1 or 3 μM H-NS respectively). 32P-labeled DNA (10 nM) was used for lanes denoted −RNAP; unlabeled DNA and 32P-labeled RNA formed by incorporation of [a-32P]GTP were used for lanes denoted +RNAP. (C) Representative AFM images of H-NS filaments on DNA or ECs matching the EMSA assays shown in (B). DNA or ECs with either 66 H-NS/kb or 200 H-NS/kb were diluted from 10 nM to 2 nM, immediately absorbed on APS-mica, and imaged in air. RNAP bound to DNA is indicated by white arrows. Cyan arrows indicate linear H-NS complexes (L), interwound H-NS complexes (I), circular H-NS complexes (C), or hairpin H-NS complexes (H). Graphics depicting the observed DNA topologies are shown in insets, where gray or black lines are each equivalent to one dsDNA molecule. AFM images from which these panels were cropped and additional examples are shown in Figure 1—figure supplement 2. (D) Pseudo-3D images of complexes similar to those in panel C, but lacking ECs to avoid scaling distortion. (E) Complexes were binned based on their DNA topology defined in (C). Interwound, circular, and hairpin H-NS complexes were grouped together as various forms of bridged complexes. H-NS complexes formed on template DNA are denoted −RNAP, and +RNAP denotes complexes formed on ECs. Only complexes with RNAP bound were counted in the +RNAP samples.
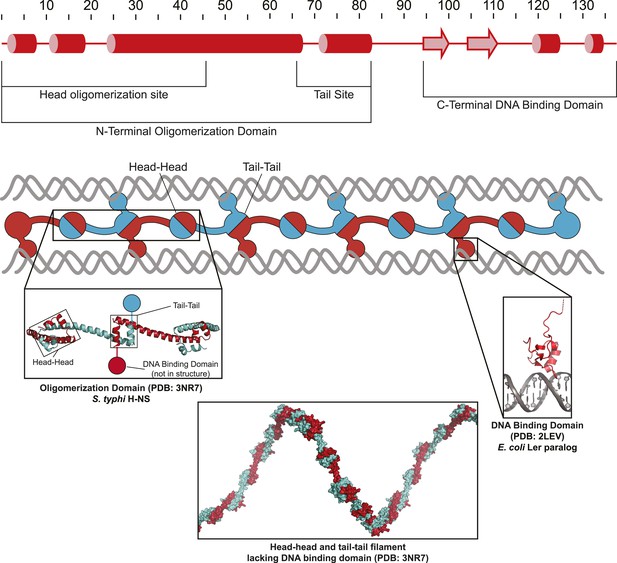
Model of H-NS filaments.
H-NS is a minor groove DNA binding protein that first binds to A/T rich DNA as a dimer through a C-terminal DNA binding domain (right inset; PDB 2LEV [Cordeiro et al., 2011]), and can form filaments by head-to-head and tail-to-tail contacts of an N-terminal oligomerization domain (left inset and bottom inset; PDB 3NR7 [Arold et al., 2010]).
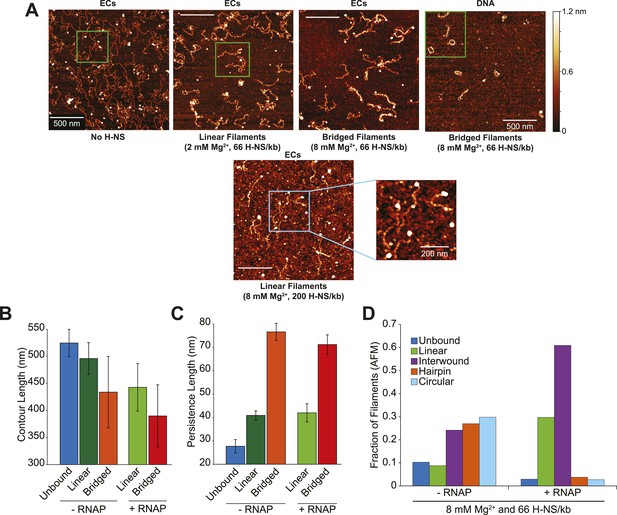
Interwound filaments formed preferentially in samples of ECs at 8 mM Mg2+ and 66 H-NS /kb.
(A) Lower magnification images of AFM as described and shown in Figure 1C, with the additional representative image of linear filaments formed in 8 mM Mg2+ and 200 H-NS/kb (high concentration H-NS). Linear filaments formed at high concentrations of H-NS have high background from the additional H-NS. Green boxes represent the area shown at higher magnification in Figure 1C. Blue box depicts the area under higher magnification in the image to the right. (B) The average contour length of H-NS filaments. Error bars represent the standard deviation of at least 25 molecules. (C) The average persistence length of H-NS filaments. Error bars represent the standard deviation of at least 25 molecules. (D) Distribution of complexes formed in bridging conditions (8 mM Mg2+ and 66 H-NS/kb) as determined by AFM.
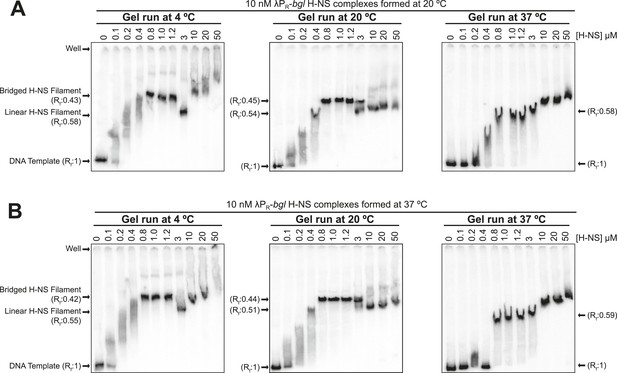
Temperature affected H-NS bridging interactions.
Native PAGE of H-NS complexes assembled at either 20°C (A) or 37°C (B) on 10 nM λPR-bgl template in 8 mM Mg2+. H-NS-DNA complexes were electrophoresed at 4°C (left panels), 20°C (middle panels), or 37°C (right panels). Rf (retardation factor) values were calculated as the distance of the H-NS-DNA complexes migrated divided by the distance the DNA template alone migrated. The loss of the ∼0.45 Rf band in gels run at 37°C but not 20°C is consistent with a loss of bridging interactions at 37°C that occurred after the samples were loaded on the gels (which were pre-equilibrated to the running temperature). Weakened H-NS interactions at 37°C also were apparent in the loss of smearing at lower H-NS concentrations (0.1–0.4 µM). The appearance at 37°C of the ∼0.55–0.58 Rf in place of the ∼0.45 Rf band evident at 20°C or 4°C suggests that linear filaments may persist at elevated temperatures. The slower migrating bands at high H-NS concentrations (≥10 µM) that appear prominently at 37°C and to some extent at lower temperatures may reflect either aggregation or restoration of bridging interactions.
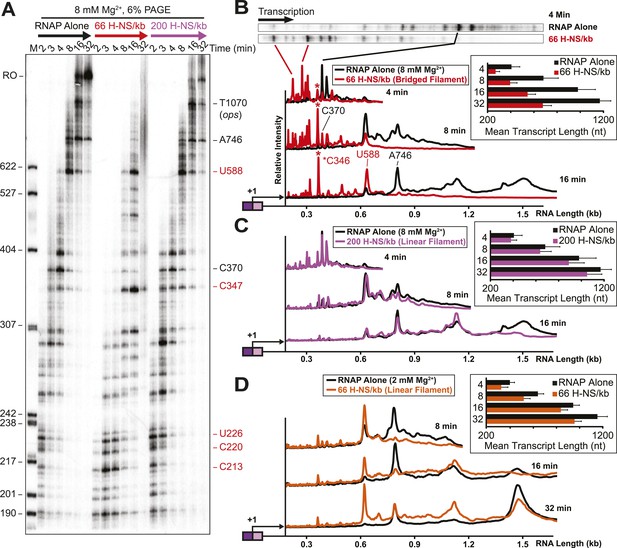
H-NS dramatically decreased transcript elongation in vitro.
(A) In vitro transcription in the presence of 66 H-NS/kb or 200 H-NS/kb filaments at 20°C, 8 mM Mg2+, and 30 µM each NTP. ECs (10 nM) were formed at the end of the C-less cassette on the λPR-bgl template (A26 ECs) and then incubated with H-NS. Samples were removed at 2, 3, 4, 8, 16, and 32 min after addition of NTPs and separated by 6% PAGE. M, 5′ end-labeled, MspI-digested pBR322 marker. RO, run-off RNA. Pauses mapped to single-nt resolution in Figure 2—figure supplement 1 and Table 1 are indicated on the right side of the gel in red for H-NS-stimulated pauses and black for H-NS independent pauses. (B, C) Densitometry profiles of transcripts produced at 8 mM Mg2+ and 20°C from the λPR-bgl template in 66 H-NS/kb or 200 H-NS/kb filaments (B and C, respectively) or without H-NS (see ‘Materials and methods’). In (B), the 4-min time point from the gel shown in (A) is displayed horizontally to allow alignment with the densitometry profile (larger transcripts are to the right). Key pauses are marked in the profiles. Insets, mean transcript lengths and standard deviations were calculated from at least four independent experiments. (D) Densitometry profiles of transcripts produced at 2 mM Mg2+ and 20°C from the λPR-bgl template in 66 H-NS monomer/kb compared to without H-NS. Inset, mean transcript lengths and standard deviations were calculated from at least four independent experiments.
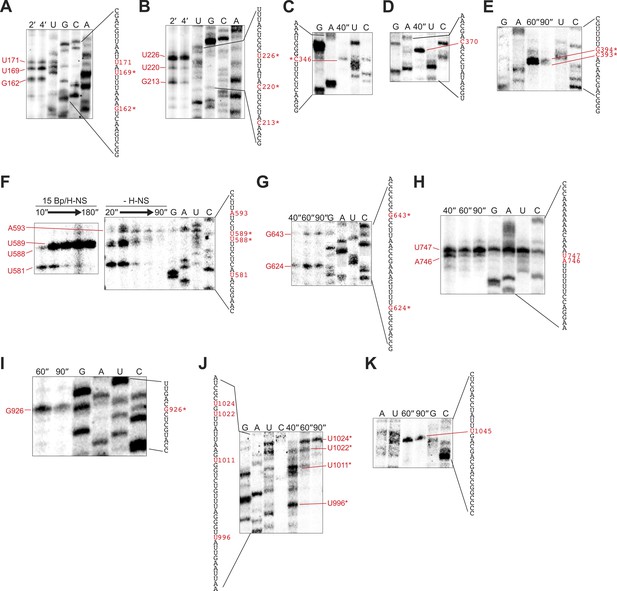
Mapping of 3′ ends of pauses on λPR-bgl template.
(A–K) Pauses on the λPR-bgl template were could be mapped to nucleotide resolution up to ∼200 nucleotides downstream from the transcription start site using ladders generated by 3′-deoxy NTP incorporation (see ‘Materials and methods’). To map pauses further downstream, we prepared truncated templates that deleted 5′ portions of the bgl transcribed region (pMK122, pMK110-520, pMK124, and pMK126; ‘Materials and methods’). Halted A26 ECs (10 nM) were formed on pMK110 template (A–B), pMK122 template (C–E), pMK110-520 template (F–G), pMK124 template (H), or pMK126 template (I–K). Transcription was then restarted with 30 μM ATP, UTP, GTP, and CTP at 37°C with or without 10–50 μM 3′-deoxy GTP, ATP, UTP, or CTP (concentrations were adjusted depending on the segment to be examined). Samples were collected at times indicated on the panels and then separated by 8% PAGE. Lanes are marked with sample times or the 3′-deoxyNTP used. Contrast in each image was adjusted to increase visibility. The mapped sequence is indicated next to each panel with the pause 3′ nucleotide highlighted in red. H-NS-stimulated pause sequences are starred. In one case (G), transcription was restarted in the presence or absence of 66 H-NS/kb at 20°C to determine which pause was H-NS dependent.
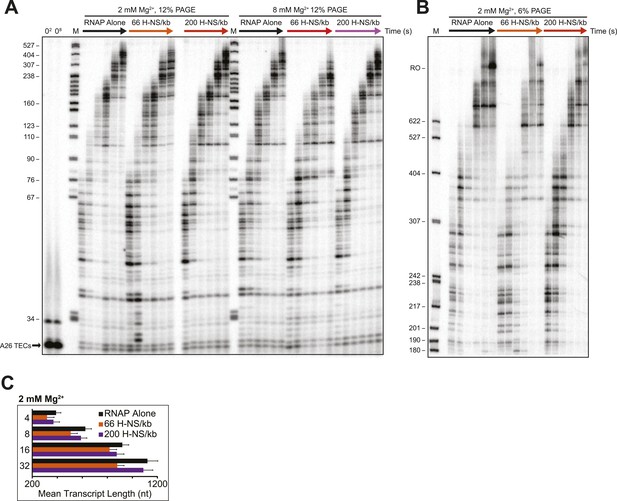
Linear H-NS filaments had minimal effects on elongation.
(A) 10 nM halted ECs formed on the λPR-bgl template were incubated with H-NS in either 2 or 8 mM Mg2+ to reach equilibrium. 30 μM NTPs were added, time points were taken at 10, 20, 40, 60, 120, and 180 s at 20°C, then resolved by 12% PAGE. M denotes labeled MspI-digested pBR322 marker. 02 and 08 refers to the time point taken prior to the addition of NTPs in 2 or 8 mM Mg2+ respectively. (B) PAGE (6% PA) of the 2 mM Mg2+ reactions described in (A). Time points were taken at 2, 3, 4, 8, 16, and 32 min at 20°C. M denotes labeled MspI-digested pBR322 marker, and RO indicates template run-off products. (C) Mean transcript lengths at various time points plotted with error bars depicting standard deviations of at least four independent experiments assembled in 2 mM Mg2+ buffer.
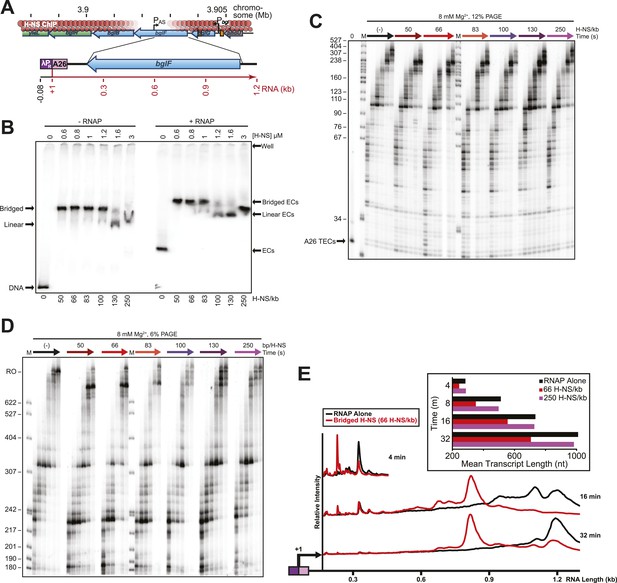
H-NS effects on transcript elongation also occurred on a different template.
(A) The 1.27-kb linear pMK121 DNA template (λPR-bglF template) similarly contains the λPR promoter followed by a 26-nucleotide C-less cassette and includes a different portion of the bgl operon, the region downstream of the bglF antisense promoter (PAS) (Peters et al., 2012). (B) Native PAGE of filaments formed on 10 nM labeled λPR-bglF template at increasing H-NS concentrations and 8 mM Mg2+ (−RNAP lanes). (+RNAP lanes), 10 nM halted A26 ECs at increasing H-NS concentrations and 8 mM Mg2+. (C) A26 ECs (10 nM; λPR-bglF template) were incubated with H-NS in 8 mM Mg2+. NTPs (30 µM) were added, samples were removed at 10, 20, 40, 60,120, and 180 s and 20°C, and the samples were resolved by denaturing PAGE (12% PA). M, labeled MspI-digested pBR322 marker. 0, time point taken prior to the addition of NTPs. (D) PAGE in 6% PA of the reactions described in (C). Samples were taken at 2, 3, 4, 8, 16, and 32 min, M, labeled MspI-digested pBR322 marker, RO, indicates template run-off products. (E) Densitometry profiles of various time points of reactions with 66 H-NS/kb compared to the absence of H-NS. Mean transcript lengths at various time points from two independent experiments are shown in the chart.
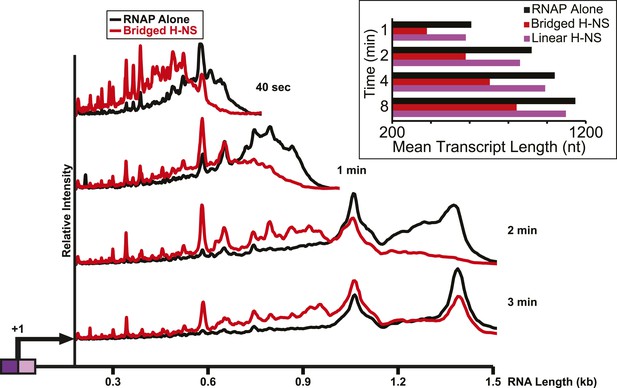
H-NS inhibited transcript elongation at physiological NTP concentrations (1 mM each NTP).
Densitometry profiles of transcripts produced at 20°C, 12 mM Mg2+ and 1 mM each NTP from the λPR-bgl template in bridged H-NS filaments (66 H-NS/kb) or in linear H-NS filaments (200 H-NS/kb). Samples were removed at 0.66, 1, 2, 3, 4, 8, and 16 min after addition of NTPs and separated by denaturing PAGE. Inset, mean transcript lengths at various times were averaged from two independent experiments.
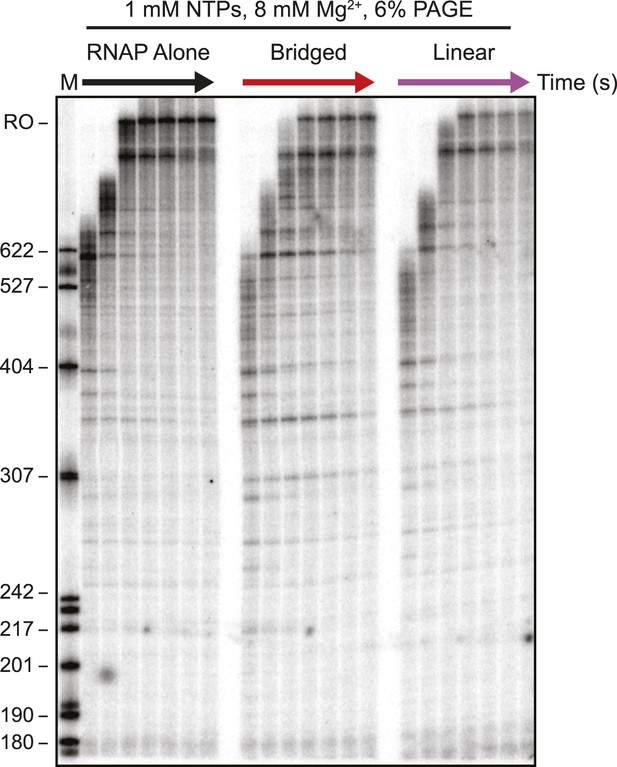
Electrophoretic gel image showing H-NS inhibited transcription elongation at physiological NTP concentrations (1 mM each NTP).
ECs (10 nM) formed on the λPR-bgl template were incubated with bridged H-NS (66 H-NS/kb) or linear filaments (200 H-NS/kb) in 12 mM Mg2+. 1 mM NTPs were added, and samples were removed at 0.66, 1, 2, 3, 4, 8, and 16 min and resolved by denaturing PAGE. M denotes labeled MspI-digested pBR322 plasmid marker, and RO indicates template run-off products.
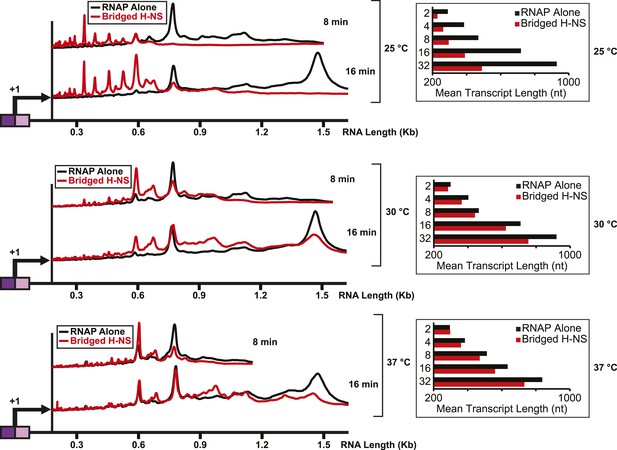
H-NS effects on transcript elongation were reduced at ≥30°C.
Densitometry profiles of transcripts produced at 25°C, 30°C, or 37°C, 8 mM Mg2+ and 30 μM each NTP from the λPR-bgl template in the presence of 66 H-NS/kb (bridged filaments). Samples were removed at 2, 4, 8, 16, and 32 min after addition of NTPs and separated by denaturing PAGE. Insets, mean transcript lengths at various time points plotted were averaged from two independent experiments.
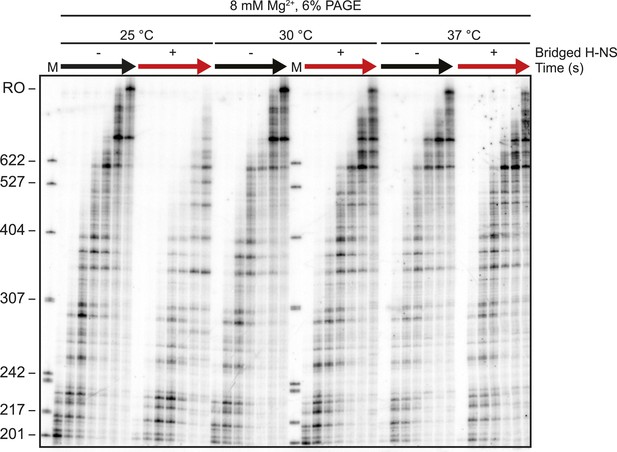
Electrophoretic gel image showing reduced H-NS effects on transcription elongation at ≥30°C.
10 nM-halted ECs formed on the λPR-bgl template were incubated with 66 H-NS/kb in 8 mM Mg2+ for 20 min at 25°C, 30°C, or 37°C. 30 μM NTPs were added, time points were taken at 2, 3, 4, 8, 16, and 32 min and resolved by PAGE. M denotes labeled MspI-digested pBR322 marker. RO indicates template run-off.
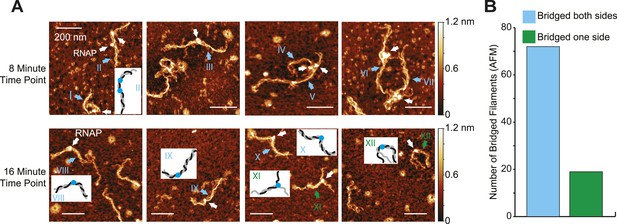
Bridged H-NS filaments reformed upstream of ECs during transcription.
(A) Representative AFM images of ECs elongating through bridged filaments (66 H-NS/kb; 8 mM Mg2+; 20°C) sampled at either 8 or 16 min after addition of NTPs (30 µM each). ECs (10 nM) and H-NS filaments were absorbed onto APS-mica and imaged in air. ECs are indicated by white arrows. Roman numerals and arrows depict two classes of filaments formed during transcription (cyan, bridged on both sides of the EC; green, unbridged on one side of the EC). Depictions of ECs and filaments are shown in insets for a subset of panels (black, gray different DNA duplexes; blue, RNAP). (B) Quantification of H-NS filament disposition during EC elongation from AFM images like those shown in (A). Cyan bar, bridged H-NS filaments both upstream and downstream of ECs. Green bar, bridged H-NS filaments on only one side of ECs.
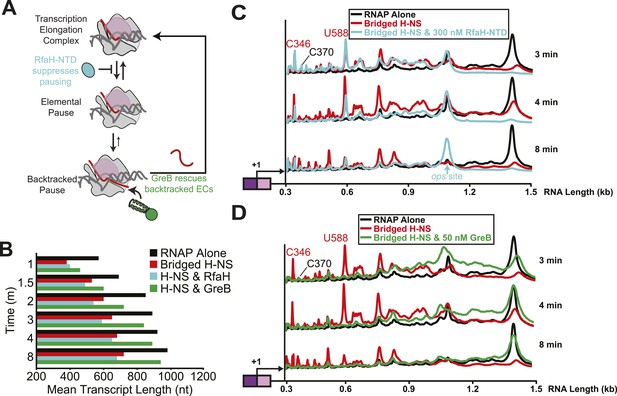
Bridged H-NS filaments induced RNAP backtracking, which was rescued by GreB.
(A) Steps in pausing affected by the NusG-like N-terminal domain (NGN) of RfaH (RfaH-NTD) or Gre factors (e.g., GreB). Binding of the NGN RfaH-NTD (cyan) to the clamp domain (pink) of RNAP inhibits clamp motion and suppresses entry into pause states (Sevostyanova et al., 2011). The duration of pausing once paused ECs form can be increased by backtracking of DNA and RNA through RNAP, during which the 3′ RNA enters the RNAP secondary channel. GreB promotes endonucleolytic cleavage of the backtracked RNA in the RNAP active site to convert an offline paused EC back to an active EC (Laptenko et al., 2003). (B) Mean transcript lengths were averaged from two independent experiments. (C, D) Densitometry profiles of transcripts produced at 20°C, 12 mM Mg2+, and 1 mM each NTP from the λPR-bgl template in bridged filaments (66 H-NS/kb) with or without 300 nM RfaH-NTD (C) or 50 nM GreB (D). Samples were removed at 0.33, 0.66, 1, 1.5, 2, 3, 4, 8, and 16 min after addition of NTPs and separated by denaturing PAGE.
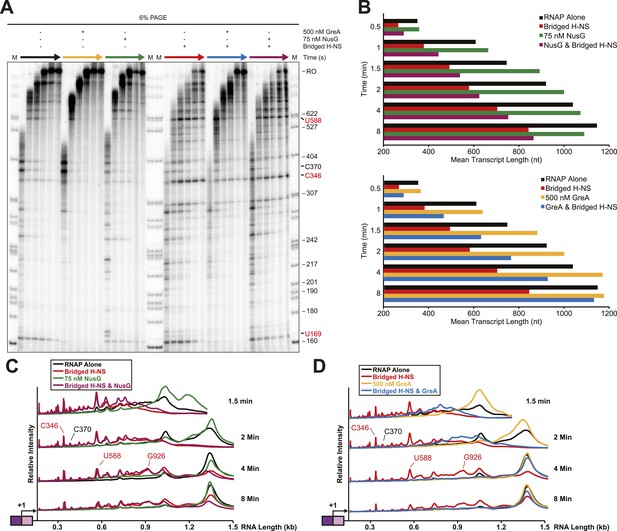
At 1 mM NTPs, NusG partially suppressed H-NS effects on pausing and GreA more significantly suppressed H-NS effects.
(A) A26 ECs (10 nM) formed on λPR-bgl were incubated with H-NS to form bridged filaments at 66 H-NS/kb and 12 mM Mg2+. GreA (500 nM) or NusG (75 nM) was added followed by NTPs (1 mM each), time points were taken at 0.5, 1, 1.5, 2, 4, and 8 min at 20°C, and separated by PAGE. M, labeled MspI-digested pBR322 marker. RO, run-off RNAs. (B) Mean transcript lengths were calculated from two independent experiments. (C, D) Densitometry profiles of transcripts produced from the λPR-bgl templates with or without bridged H-NS and with or without GreA (C) or NusG (D).
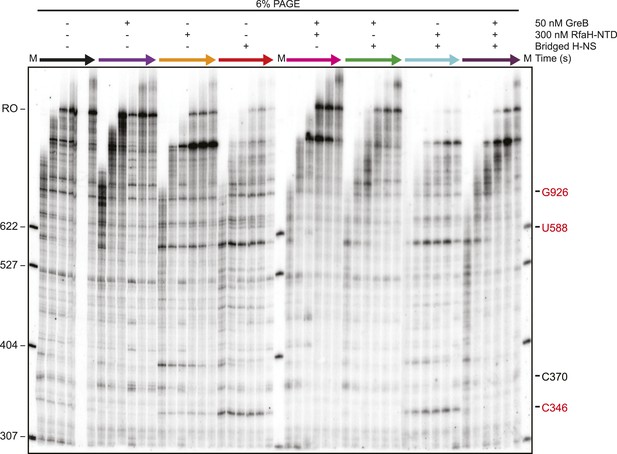
Electrophoretic gel image showing that H-NS-induced RNAP backtracking was rescued by GreB.
PAGE corresponding to Figure 6, where reactions were assembled with 66 H-NS/kb and 50 nM GreB or 300 nM RfaH-NTD.
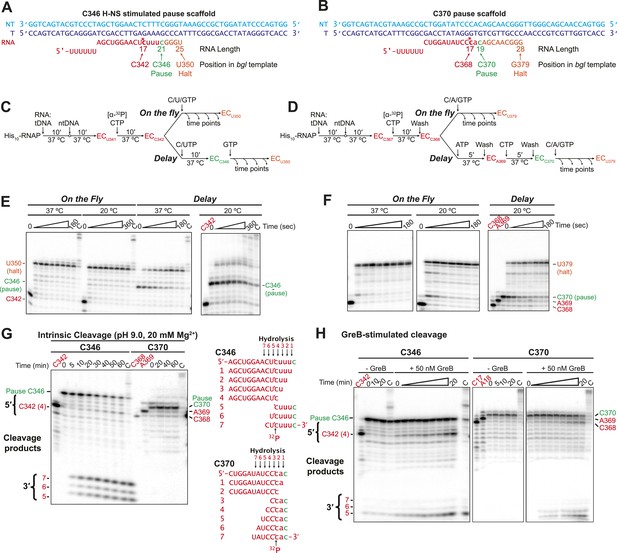
The H-NS stimulated C346 pause readily backtracked, whereas H-NS-resistant pausing at C370 occurred without obligate backtracking.
(A, B) Nucleic-acid scaffolds that enable EC reconstitution just upstream of sequences equivalent to the C346 H-NS-stimulated pause (A) and the C370 pause not stimulated by H-NS (B). Lowercase RNA, nt added by RNAP after reconstitution. *, position of [32P]CMP incorporation (red). Green, 3′ nt at the pause. (C, D) Pause assay reaction schemes (see ‘Materials and methods’). For assays on the fly, 10 μM CTP, UTP, and GTP (for ECC346) or CTP, ATP, and GTP (for ECC370) were added to ECs upstream from the pause at either 37°C or 20°C. For C346 delay assay (C), 10 μM CTP and UTP extended ECs to the pause and 10 μM GTP was added after 5 min to extend the RNA. For C370 delay assay (D), ECs were immobilized on Co2+ magnetic beads and extended to the pause by stepwise incubation with [α-32P] CTP, ATP, and CTP, incubated for 5 min, and then extended from the pause with CTP, ATP, and GTP (all at 10 μM NTP). ECs were washed five times with 1 ml EB between steps. (E, F) Denaturing RNA gels of products of assays depicted in (C) and (D). C, chase sample incubated with 1 mM all 4 NTPs. (G, H) C346 or C370 paused ECs formed as shown in (C) and (D) were resuspended in cleavage buffer (pH 9.0 and 20 mM Mg2+) to induce intrinsic cleavage (G) or in EB with or without 50 nM GreB to induce GreB-mediated hydrolysis (H). Possible 5′ and 3′ cleavage products determined by position of label are illustrated for C346 and C370 between the gel panels.
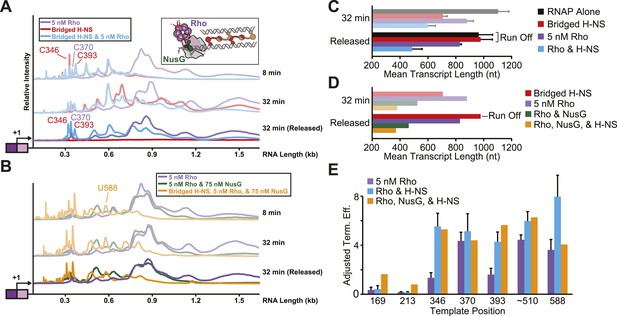
Stimulation of pausing by bridged H-NS filaments aided Rho-dependent termination.
(A, B) Densitometry profiles of transcripts produced at 28°C, 8 mM Mg2+, and 30 µM each NTP from the λPR-bgl template in bridged filaments (66 H-NS/kb) with or without 5 nM Rho (A) and with or without 75 nM NusG (B). Samples were removed at 2, 4, 8, 16, and 32 min after NTPs were added and separated by denaturing PAGE. To detect release of Rho-terminated transcripts, a 32-min sample was separated into released and EC-bound fractions using paramagnetic Co2+ beads that bind the His10-tagged RNAP. The released supernatant fraction was separated by denaturing PAGE and converted to densitometric profiles shown as darker colors. (C, D) Mean transcript lengths and standard deviations were calculated from at least two (C) or four (D) independent experiments. (E) Rho termination efficiencies were calculated as the fraction of released transcripts divided by the total transcripts, with averages and standard deviations from at least three independent experiments.
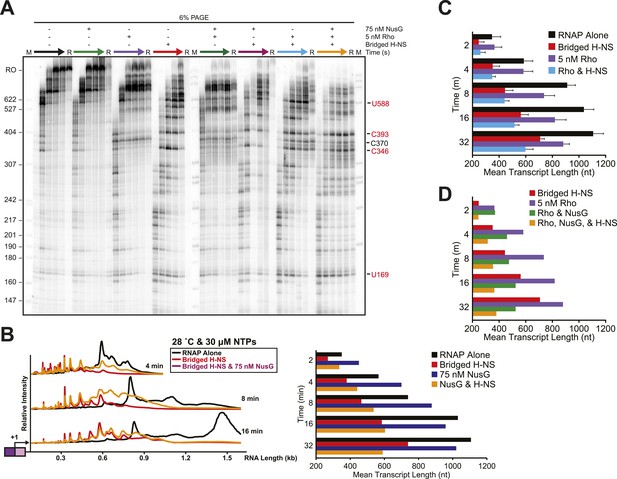
Stimulation of pausing by bridged H-NS filaments aided Rho-dependent termination.
(A) Halted A26 ECs (10 nM) formed on the λPR-bgl template were incubated with bridged H-NS, 75 nM NusG, or 5 nM Rho at 8 mM Mg2+ at 28°C. Samples were then collected at 2, 4, 8, 16, and 32 min after addition of NTPs (30 µM each) and resolved by PAGE. Rho-terminated, released transcripts for the 32-min sample were determined as described in the legend to Figure 7. R, released transcripts. M, labeled MspI-digested pBR322 marker. RO, run-off RNAs. (B) Densitometry profiles were determined as described in legend of Figure 2. Mean transcript lengths were averaged from at least two independent experiments. (C) Mean transcript lengths and standard deviations with or without bridged H-NS and with or without 5 nM Rho were determined using at least four independent experiments. (D) Mean transcript lengths with or without bridged H-NS, 5 nM Rho, or 75 nM NusG were averaged from two independent experiments.
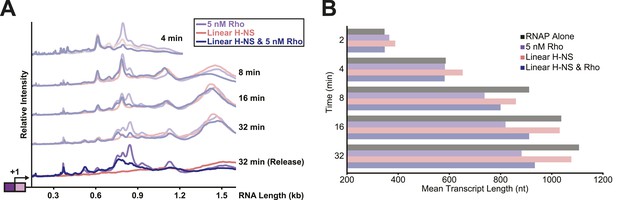
Linear H-NS filaments did not aid in Rho-dependent termination.
(A) Densitometry profiles of transcripts produced at 28°C, 8 mM Mg2+, and 30 µM each NTP from the λPR-bgl template in linear filaments (200 H-NS/kb) with or without 5 nM Rho. Samples were removed at 2, 4, 8, 16, and 32 min after addition of NTPs and separated by denaturing PAGE. Rho-terminated, released transcripts for the 32-min sample were determined as described in the legend to Figure 7. (B) Mean transcript lengths were averaged from two independent experiments.
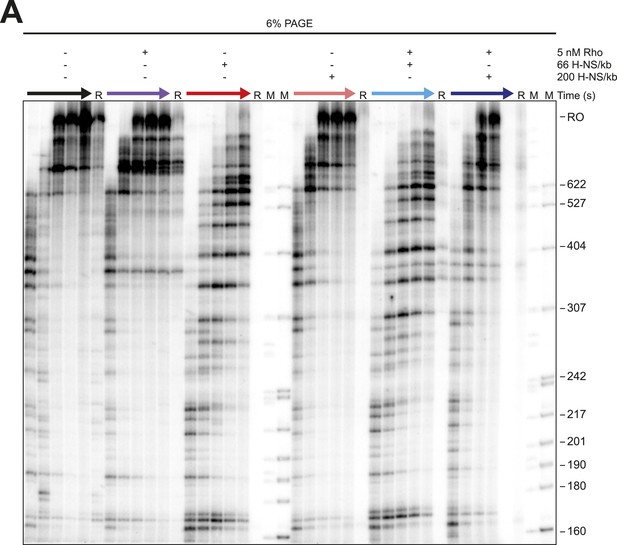
Linear H-NS Filaments did not aid in Rho termination.
(A) Halted A26 ECs (10 nM) formed on λPR-bgl template were incubated with either 66 H-NS/kb or 200 H-NS/kb with or without 5 nM Rho at 8 mM Mg2+ and 28°C. Samples were then collected at 2, 4, 8, 16, and 32 min after addition of NTPs (30 µM each) and resolved by PAGE. Rho-terminated, released transcripts for the 32-min sample were determined as described in the legend to Figure 7. R, released RNAs, M, labeled MspI-digested pBR322 marker. RO, run-off RNAs.
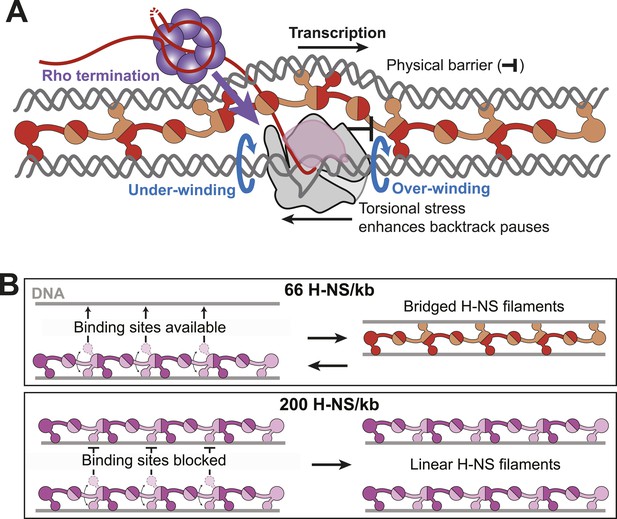
Models for H-NS effects on pausing, Rho termination, and DNA bridging.
(A) As RNAP elongates through bridged filaments, pause durations increase for one or both of two reasons: (i) the off-rate of bridged H-NS is slower than the elongation rate of RNAP, leading to a roadblock (physical barrier; black bar); or (ii) H-NS bridging creates a closed topological domain that accumulates positive and negative supercoiling (torsional stress) in front and behind the EC, respectively, because free rotation of the DNA is blocked by bridged H-NS contacts and free rotation of the EC is blocked by steric clash between the bridged H-NS–DNA filament and the nascent RNA, including macromolecules like Rho or ribosomes bound to the nascent RNA (Liu and Wang, 1987). Blue arrows depict the rotation of DNA required to avoid torsional strain when the DNA is unconstrained. Both the under-winding (behind EC) and over-winding (in front of EC) torsional stresses will increase the propensity for RNAP to backtrack, thus increasing the duration of pauses that involve backtracking and increasing the kinetic window for Rho-dependent termination at backtrack pauses. (B) At 66 H-NS/kb, H-NS-free DNA segments allow DNA-binding domains from initially formed linear filaments to interact and form bridged filaments (top). At 200 H-NS/kb, all DNA segments become occupied by H-NS, leaving no available unbound DNA for formation of bridged filaments (bottom).
Tables
Pause sites and their responses to H-NS and transcription factors
| Pause position | Pausing | Termination | Sequence | ||||||
|---|---|---|---|---|---|---|---|---|---|
| H-NS | RfaH | GreB | GreA | Rho | Rho + H-NS | Rho + NusG | Pause↓ | ||
| C134 | ↑ | ? | ? | ? | ? | ? | ? | CGCUGAUAACUCAAGC | UUUCUUCCUG |
| G162 | ↑ | ↓ | ↓ | ? | – | – | – | AAUUAAGGCUGAACUG | AAAUUUUAUU |
| U169 | ↑ | – | ↓ | ↓ | – | – | ↑ | GCUGAACUGAAAUUUU | AUUAAUUGCA |
| C213 | ↑ | – | ↓ | ↓ | – | – | – | GCGUGACACCUGCAAC | AUCCUCCAUA |
| C220 | ↑ | ↓ | – | ↓ | – | – | – | ACCUGCAACAUCCUCC | AUAUUUCCGC |
| U226 | ↑ | ↓ | – | ↓ | – | – | – | AACAUCCUCCAUAUUU | CCGCUCAUUU |
| C346 | ↑↑↑ | – | ↓ | – | – | ↑ | – | UAGCUGGAACUCUUUC | GGGUAAAGCC |
| C370 | – | ↓ | – | ↓ | ↑ | – | – | CCGCUGGAUAUCCCAC | AGCAACGGGU |
| C393, G394 | ↑ | ↓ | ↓ | ↓ | – | ↑ | ↑ | GGUUGGGCAGCAACAC | GUUUUGCUGA |
| U588, U589 | ↑↑↑ | – | ↓ | ↓ | ↑ | – | – | UCAAGGCAUACUCUUU | UUCUAUUCCA |
| A593 | – | – | – | – | – | – | – | GCAUACUCUUUUUCUA | UUCCACUUGA |
| G624 | ↑ | ↓ | ↓ | ↓ | – | – | – | UUCUUUCGCCAGCGCG | UUUUUGAAAG |
| G643 | ↑ | – | ↓ | ↓ | – | – | – | UUGAAAGCCAAUUCCG | CGCCCCAUGA |
| A746, U747 | – | – | ↓ | – | ↑ | – | ↑ | GCAAGGACCUUUUUUA | UAAACAAAAA |
| G926 | ↑ | – | – | – | – | ? | ? | AAUAUGACCAUGCUCG | CAGUUAUUAA |
| U996 | ↑ | – | ↓ | ↓ | – | ? | ? | CCAAUAAUUAAGUUAU | UGGGAUUUGU |
| U1011 | ↑ | ↓ | ↓ | ↓ | – | ? | ? | UUGGGAUUUGUCUGGU | GAAUUAUUUG |
| U1022, U1024 | ↑ | ↓ | ↓ | ↓ | – | ? | ? | GUCUGGUGAAUUAUUU | GUCGCUAUCU |
| U1079 (ops) | – | – | ↓ | ↓ | ↑ | ? | ? | CUAGUGGCGGUAGCGU | GCUUUUUUCA |
-
Pause positions are given as 3′ RNA nucleotide identity and distance from the transcription start site as mapped by high-resolution PAGE (Figure 2—figure supplement 1). ↑, increased pause or termination. ↓, decreased pause or termination. In the sequences shown, pause 3′ ends are bold (under arrow) and the position corresponding to the incoming NTP is underlined.





