Synthetic enzyme-substrate tethering obviates the Tolloid-ECM interaction during Drosophila BMP gradient formation
Figures
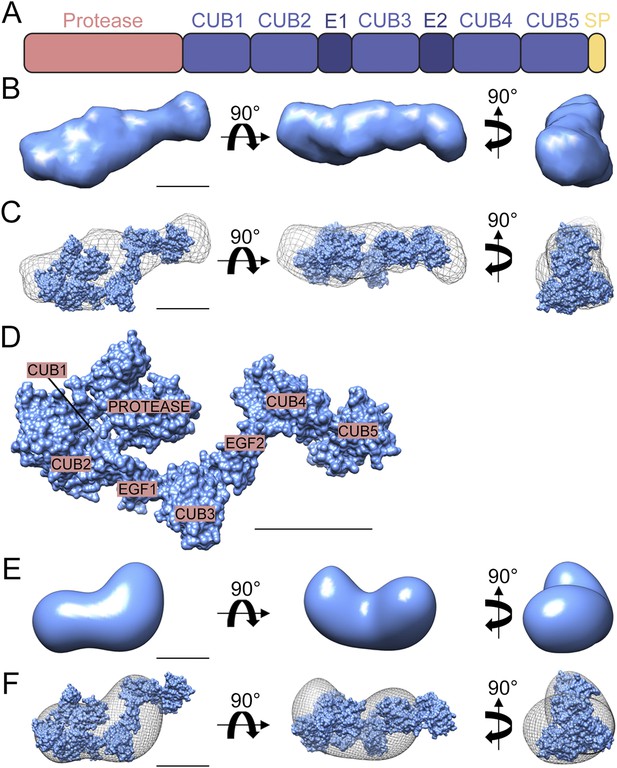
Structure of Drosophila Tld.
(A) Cartoon of Drosophila Tld domain organisation. E = EGF-like domain. SP = specific peptide. (B) Low resolution ab initio model generated by DAMMIN and DAMAVER from solution SAXS data shown in three orthogonal orientations. (C) Overlay of the ab initio model shown in (B) displayed as chicken-wire with a representative rigid body model generated by SASREF. (D) The rigid body model shown in (C) with Tld domains labelled. (E) 3D reconstruction of the Drosophila Tld monomer by single particle analysis electron microscopy shown in three orthogonal orientations. (F) The rigid body model was fitted into the EM volume shown in (E), here displayed as chicken-wire. All scale bars = 5 nm. See also Figure 1—figure supplements 1 and 2.
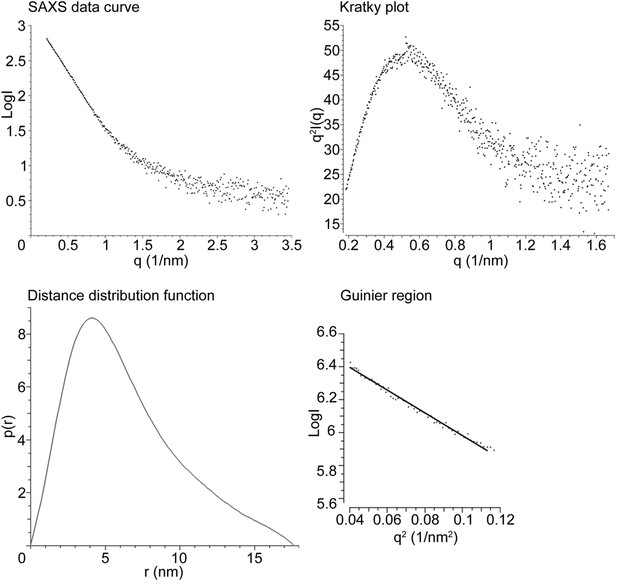
Drosophila Tld SAXS data analysis.
Drosophila Tld SAXS data showing the SAXS data curve, Kratky plot, Distance distribution function (p(r)) and Guinier plot. Maximum particle dimension is indicated by the point at which the p(r) function reaches zero.
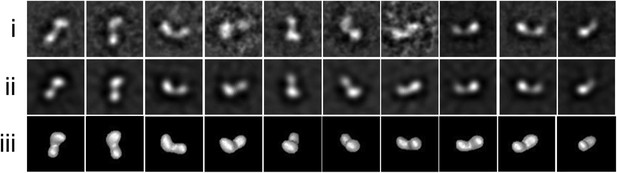
Drosophila Tld TEM data.
Single particle EM data (i) Representative class averages used in the 3D reconstruction after 10 iterations of multi-reference alignment and refinement box size shown. (ii) Re-projections from the Drosophila Tld 3D EM model. (iii) Surface views of the Tld 3D reconstruction as calculated by angular reconstitution. All box sizes = 23 × 23 nm.
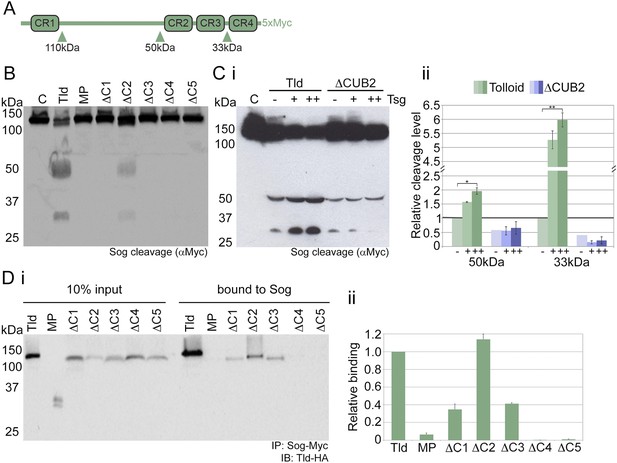
Tld CUB4/5 domains mediate Sog binding.
(A) Schematic of Sog showing the CR domains and the position of the Tld cleavage sites. The sizes of the C-terminal fragments liberated by cleavage, as detected by the C-terminal Myc tag, are indicated below the arrows. (B) Western blot (anti-Myc) of Sog cleavage assays carried out using normalised amounts of the Tld deletion proteins indicated, all in the presence of Dpp and absence of Tsg. C = control assay with no Tld added. MP = the isolated metalloproteinase domain. (Ci) A representative Western blot (anti-Myc) showing Sog cleavage assays carried out with normalised levels of Tld and ΔCUB2 in the presence of Dpp and either in the absence (−), or with increasing amounts (+, ++) of Tsg. (Cii) Graph shows the amount of cleavage measured as the amount of the 50 kDa and 33 kDa fragments quantified relative to cleavage calculated for wildtype Tld in the absence of Tsg, based on three different experiments including the Western blot shown in i. Error bars show SEM, n = 3, *p < 0.005, **p < 0.001. (Di) Western blot (anti-HA) showing 10% input of catalytically inactive forms of Tld-HA proteins and the amount bound to Sog-Myc by immunoprecipitation. (Dii) Quantitation of the level of binding of each Tld protein to Sog, relative to binding of full-length Tld. Error bars are SEM, n = 4. See also Figure 2—figure supplements 1 and 2.
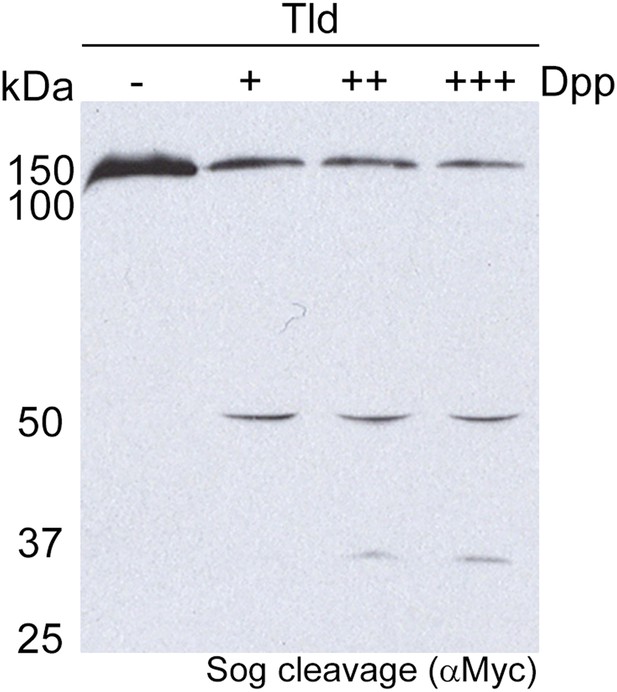
Sog cleavage by Tld is dependent on Dpp.
Western blot (anti-Myc) showing Sog cleavage by Tld in the presence of increasing amounts (+, ++, +++) of the Dpp ligand. No Sog cleavage by Tld is detectable in the absence of Dpp.
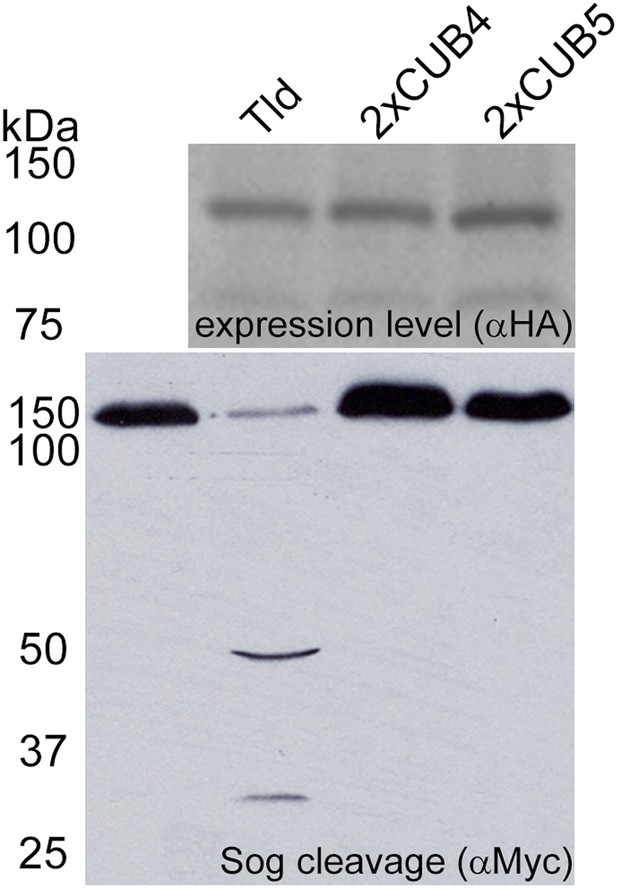
Tld CUB4-CUB5 domain pair is specifically required at the C-terminus.
Western blot (anti-HA) of the indicated Tld protein levels following expression in Drosophila S2R+ cells with Sog cleavage in the presence of Dpp shown below on an anti-Myc Western blot. The 2xCUB4 and 2xCUB5 variants retain five CUB domains but have a CUB4 or CUB5 duplication at the C-terminus instead of the CUB4-5 domain pair.
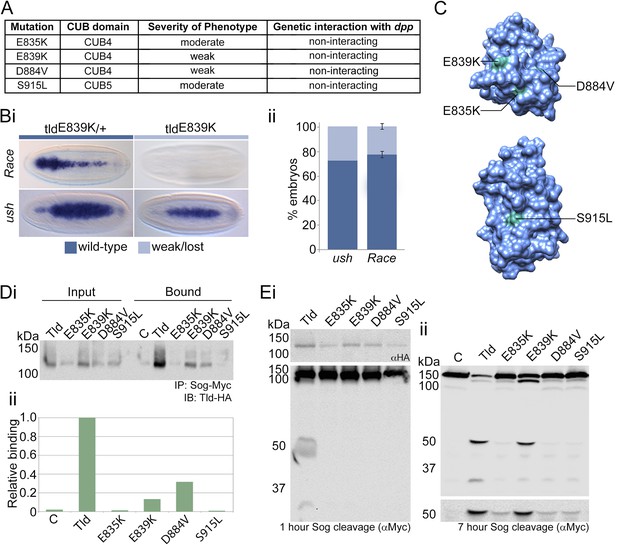
Altered Sog binding of Tld point mutants.
(A) Table summarising previous experimental data regarding the CUB4 and CUB5 mutations and the severity of the resulting phenotypes (Childs and O'Connor, 1994; Finelli et al., 1994). (Bi) Dorsal views of stage 6 wildtype and tldE839K homozygous embryos stained by RNA in situ hybridisation for the Dpp target genes Race and u-shaped. Phenotypes were quantified in Bii as the percentage of embryos with either wildtype or lost/narrow expression for Race and ush. (C) Models of Tld CUB4 and CUB5 with the single amino acid mutations highlighted in green. (Di) Western blot with anti-HA showing the input levels of the catalytically inactive Tld-HA proteins tested and the amounts bound to Sog-Myc in an immunoprecipitation experiment. (Dii) Graph shows the level of binding to Sog relative to that for full-length Tld. (E) Western blot (top, anti-HA) showing the levels of Tld proteins carrying the indicated single amino acid changes. Western blots (anti-Myc) show 1 hr (Ei) or 7 hr (Eii) Sog cleavage assays using the Tld proteins shown. Below the blot on the right is a longer exposure of the 50 kDa cleavage fragment.
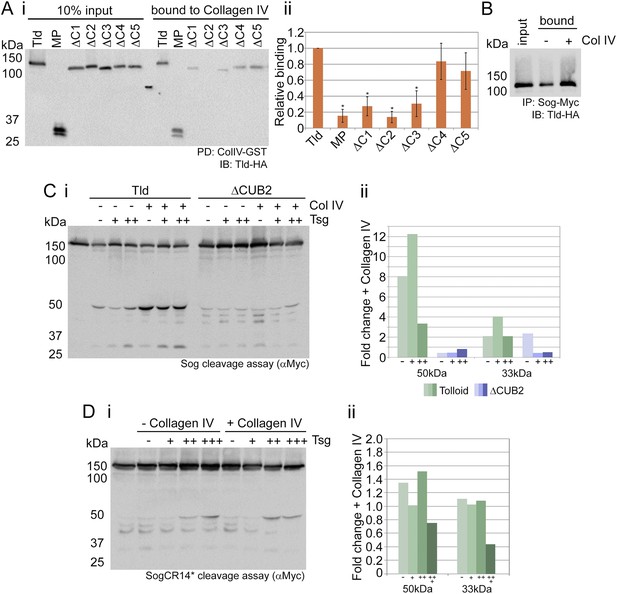
Tld binds Collagen IV.
(Ai) Western blot showing Tld-HA input proteins and the amount bound to GST-VkgNC1 in a pulldown experiment. (Aii) Graph shows the level of binding to Collagen IV plotted relative to binding of full-length Tld, based on data from three experiments. Error bars are SEM, n = 3, *p < 0.05. (B) Western blot (anti-HA) showing Tld input and the levels bound to Sog in the presence of Dpp, with or without (±) added Collagen IV. (Ci) Western blot (anti-Myc) of Sog cleavage by full-length Tld or the ΔCUB2 mutant quantified in (Cii). The x-axis labels (–,+,++) refer to the absence, or increasing amounts, of Tsg. Error bars are SEM, n = 3. (Di) Western blot (anti-Myc) of Tld cleavage of SogCR14*, with cleavage levels quantified in (Dii). Both cleavage assays were carried out over 3 hr, in the presence of Dpp, and in the absence or presence of increasing amounts of Tsg, with or without of Collagen IV, as indicated. Graphs show the fold-change in the relative levels of the 50 kDa and 33 kDa cleavage fragments released by Tld (green) or ΔCUB2 (blue), in the presence of Collagen IV relative to its absence. See also Figure 4—figure supplement 1.
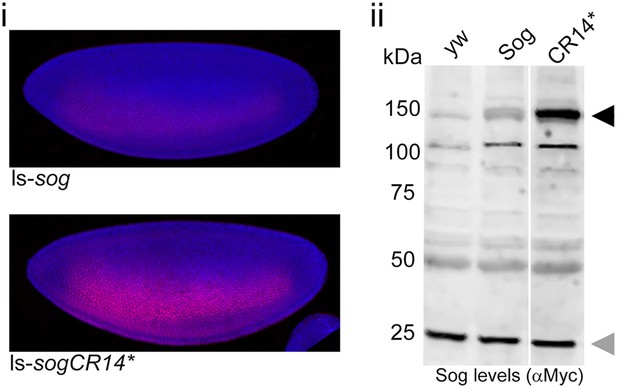
SogCR14* is inefficiently processed by Tld in vivo.
(i) Confocal projections showing lateral views of stage 5 ls-sog-Myc and ls-sogCR14*-Myc transgenic embryos stained with anti-Myc (red) and DAPI (blue). Embryos were stained in parallel and imaged with the same settings. (ii) Western blot (anti-Myc) of yw, ls-sog-Myc and ls-sogCR14*-Myc embryo extracts. All samples were analysed on the same Western blot, the cut in the blot only reflects removal of an irrelevant lane. ls-sogCR14* embryo extracts contain higher levels of unprocessed Sog at 150 kDa (black arrowhead) than ls-sog embryos. The non-specific band at ∼25 kDa marked by the grey arrowhead serves as a loading control. Both sog and sogCR14* transgenic lines were generated by attB/attP recombination into cytosite 86Fb on chromosome 3 to eliminate position effects.
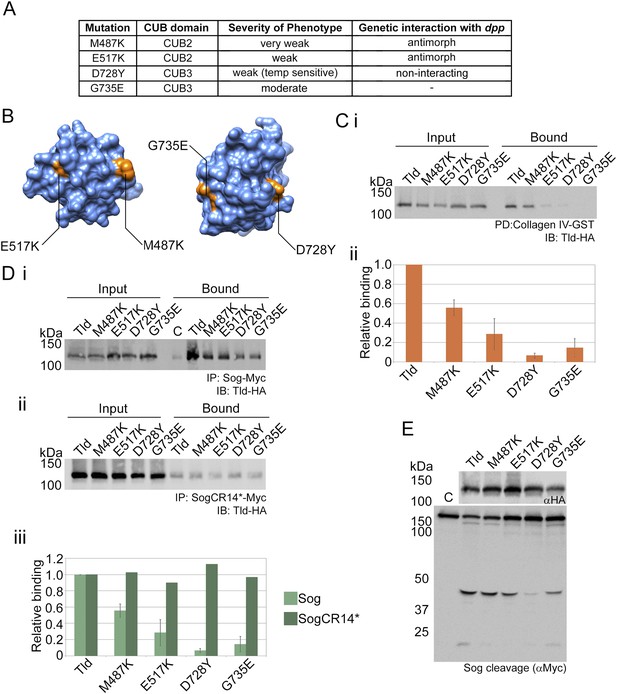
Point mutations affect Collagen IV binding.
(A) Overview of previous data relating to classical tld alleles carrying point mutations in the CUB2 and CUB3 domains detailing the single amino acid change, extent of ventralisation and genetic interaction with dpp (Childs and O'Connor 1994; Finelli et al. 1994). (B) Models of Tld CUB2 and CUB3 with the single amino acid mutations highlighted in orange. (Ci) Western blot showing binding of the Tld-HA proteins to GST-VkgNC1, in the presence of Dpp, relative to the input levels. (Cii) Graph showing quantification of binding to Collagen IV normalised to the input levels, relative to binding of full-length Tld. Error bars are SEM, n = 3. (D) Western blot with anti-HA showing input levels of the catalytically inactive Tld-HA proteins tested and the amounts bound to Sog-Myc (Di) and SogCR14*-Myc (Dii) in immunoprecipitation experiments. (Diii) Graph showing the level of binding to Sog (light green) and SogCR14* (dark green) relative to that for full-length Tld in each case. Error bars for Sog are SEM, n = 3. (E) Western blot (top, anti-HA) showing Tld-HA proteins with the indicated single amino acid changes. Western blot (lower, anti-Myc) showing Sog cleavage assays (4 hr) using the Tld enzyme indicated above each lane in the presence of Dpp.
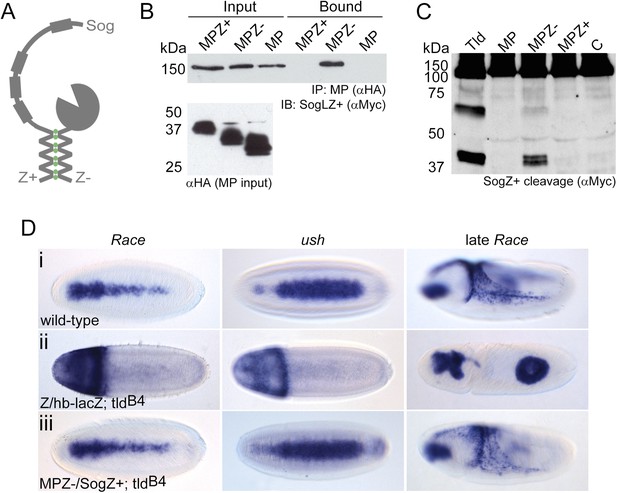
Restoration of in vivo function to the Tld MP domain.
(A) Cartoon of the leucine zipper strategy. (B) Western blot (anti-HA) showing the input levels of the MP-HA and MP-Zipper-HA fusion proteins tested, and binding of Sog-Zipper+-Myc following immunoprecipitation with anti-HA. MPZ+ is larger than MPZ− due to the presence of additional glycine residues preceding the zipper sequence. (C) Western blot (anti-Myc) showing cleavage assays carried out with SogZ+ transfected with the indicated Tld MP-zipper fusion proteins in the presence of Dpp. C = control assay without Tld added. (D) Dorsal views of stage 6 embryos, oriented with anterior to the left, stained by RNA in situ hybridisation for the Dpp target genes Race and u-shaped, and stage 9 lateral views of late Race expression. Wild-type embryos are shown for reference in (Di). Embryos in (Dii) and (Diii) are also stained with a lacZ RNA probe that marks the chromosome carrying wildtype tld (ftz-lacZ, stripes, not shown) or chromosome lacking the zipper transgene (hb-lacZ, anterior staining). All embryos shown are tld mutant, and either lack at least one zipper transgene (Dii), or carry both zipper transgenes (Diii). See also Figure 6—figure supplement 1.
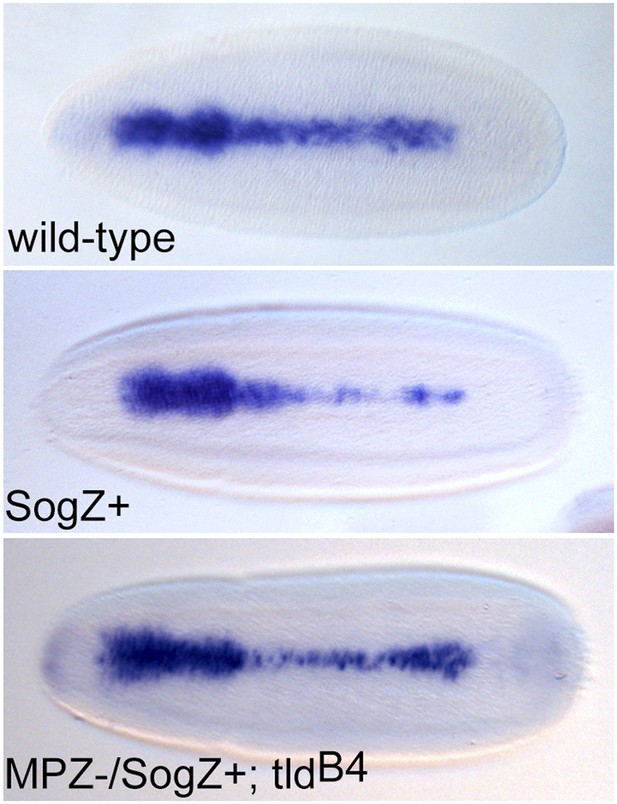
SogZ + overexpression results in a thinning of Race expression.
Dorsal views of stage 6 embryos stained for Race expression by RNA in situ hybridisation. Embryos that overexpress SogZ+ in a wild-type background show a thinning of Race expression in the central region of the embryo. 20% of MPZ−/SogZ+;tldB4 embryos show a similar phenotype.
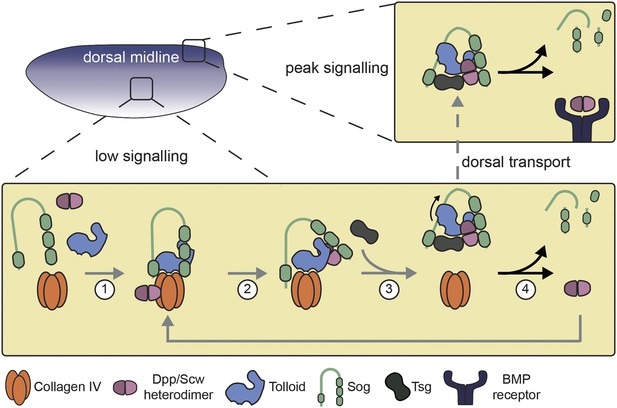
Model of Dpp gradient formation showing the Tld-ECM interaction.
See text for details.





