Ribosomes slide on lysine-encoding homopolymeric A stretches
Figures
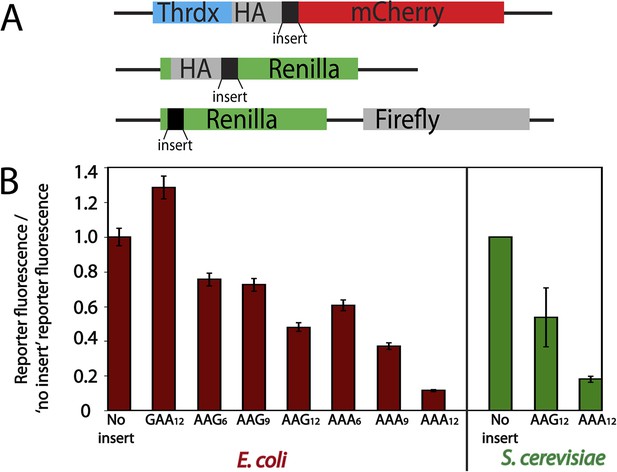
Protein production is differentially diminished by iterated lysine codons (AAA vs AAG) in E. coli and S. cerevisiae.
(A) Schematics of the mCherry (top) and luciferase (middle, and bottom) reporters used in this study. The mCherry reporter contains an N-terminal thioredoxin (Thrdx) domain, 3HA-tag, sequence of interest (black section), followed by the C-terminal mCherry sequence. The top luciferase reporter includes a 2HA tag followed by sequences of interest (used for study in Figure 1B). The second luciferase reporter (used in Figure 6) has sequences of interest at the N-terminal end of Renilla. Firefly is used in this construct as an internal control in the second luciferase reporter. (B) Relative amounts of protein expressed from reporters expressed in E. coli (mCherry, red) and S. cerevisiae (luciferase, green). Error bars results from for the standard error of at least three experiments.
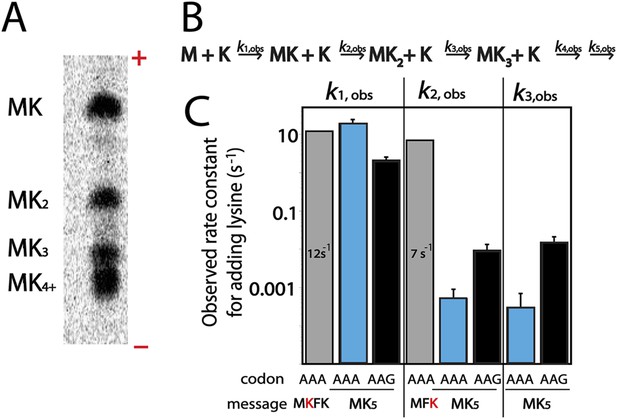
Kinetic defect observed on addition of second and third lysine residues in iterated lysine stretch.
(A) Example eTLC displaying the E. coli translation products of a AUG-(AAA)5-UAA message. The ± poles of the electrophoretic TLC are indicated. MK4 and MK5 products (and those with greater numbers of lysine) are difficult to resolve in this system but the other products are easily visualized. (B) Kinetic scheme for rate constants of sequential lysine additions to peptide chain. (C) Bar graph displaying rate constants for the addition of individual lysines to a variety of messages: MKFK-Stop (gray), MKA5-Stop (blue), MKG5-Stop (black), and MFK (gray).
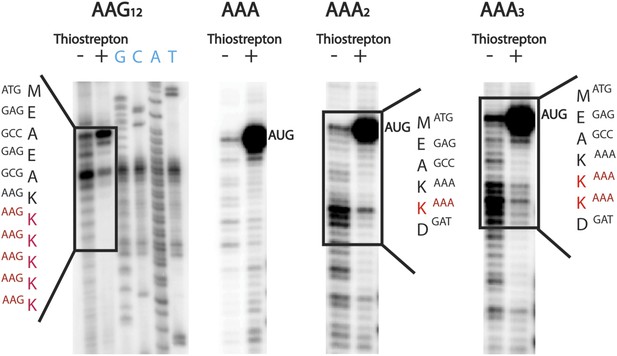
Ribosomes stall while adding a second lysine.
Toeprinting assays were performed with constructs containing 1–12 consecutive lysines inserted (either AAG and AAA codons). Assays were performed in the presence and absence of thiostrepton to mark ribosomes on the initiating AUG codon. Sequences on which toeprints appear are highlighted in red.
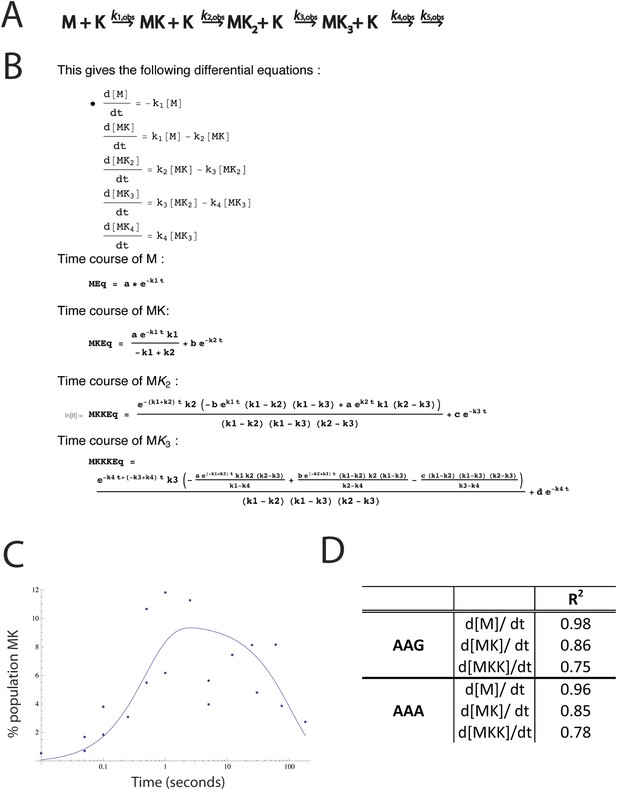
Modeling of rate constants in Mathematica.
(A) Kinetic scheme used to model the rate constants of sequential lysine additions to the peptide chain (same as Figure 2A). We also attempted to model with peptidyl-tRNA drop-off rates included. Inserting peptidyl-tRNA drop-off into our model decreases the quality of fits, and returns rates of drop-off small enough that they are inconsequential relative to the time scale of the reaction. (B) The top panel displays the differential equations used to solve for each rate constant. The bottom panels display the mathematical solutions for the differential equations. These solutions were used to perform modeling and fit the data. The fits were performed both iteratively (e.g., we solved for k1 by fitting the plots measuring the disappearance of M, then input that value into the equation describing the appearance of MK to solve for k2) and by letting all of the values float for each data set. In both cases, the rate constants modeled were essentially the same, indicating that the first lysine is added quickly (k1), and subsequent lysines (k2, k3) are added more slowly. (C) An example fit in Mathematica showing time course for the formation and depletion of MK on a message with AAG codons. This time course, for example, was used to model the k2 value. (D) R2 values for the fits for the appearance and disappearance of each species used to model rate constants.
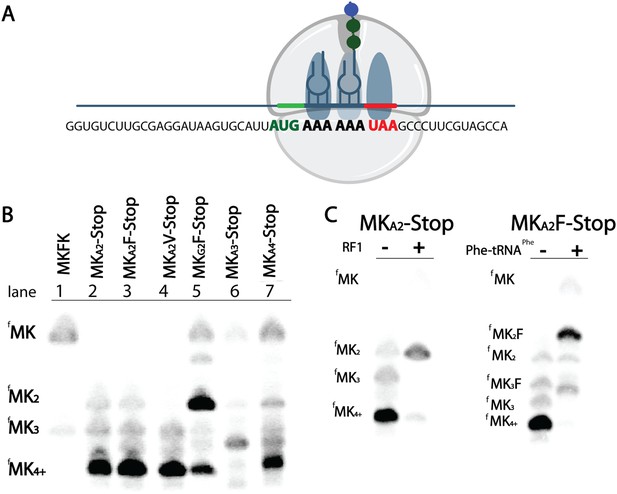
E. coli ribosomes add extra lysines on messages containing two sequential AAA, but not AAG, lysine codons.
(A) Illustration of the ribosome on the entire MKA2-Stop message. (B) eTLCs showing the peptide products resulting from translation of indicated messages with Lys-tRNAlys (but no other tRNAs or release factors) present. (C) eTLC displaying the peptide products resulting from the translation of indicated messages in the presence of Lys-tRNALys alone, or in the presence of Lys-tRNALys + factors (either RF1 or Phe-tRNAPhe) necessary for messages to be fully translated.
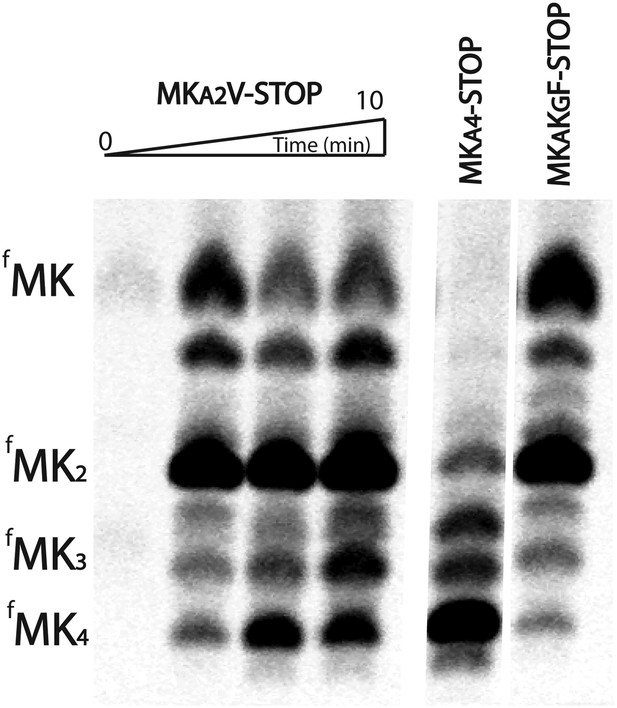
E. coli ribosomes add extra lysines to peptides translated on messages containing sequential AAA-AAG lysine codons.
TLC showing all of the peptide products resulting from translation of MKA2V-Stop, MKA3-Stop, MKA4-STOP, and MKAKGF-Stop messages with Lys-tRNAlys (but no other tRNAs or release factors) present.
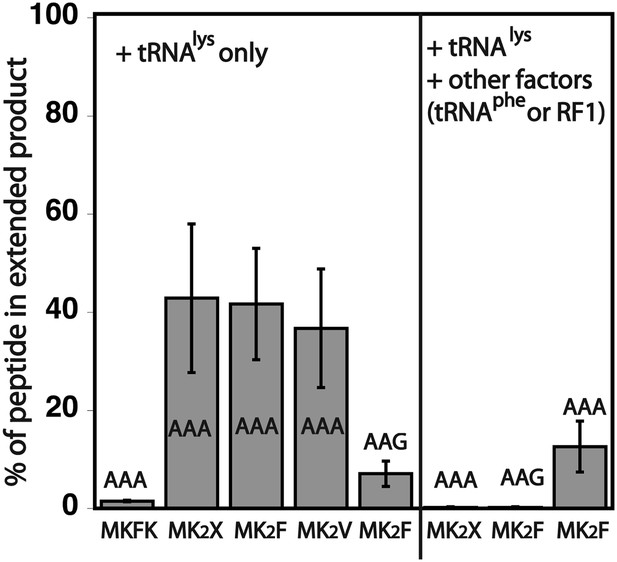
Quantification of the percentage of translated peptide containing more lysine residues than expected.
Translation reactions were run in the presence of either Lys-tRNAlys only, or Lys-tRNAlys and other factors (Phe-tRNAPhe or RF1). All errors bars represent the standard error from at least three independent experiments.
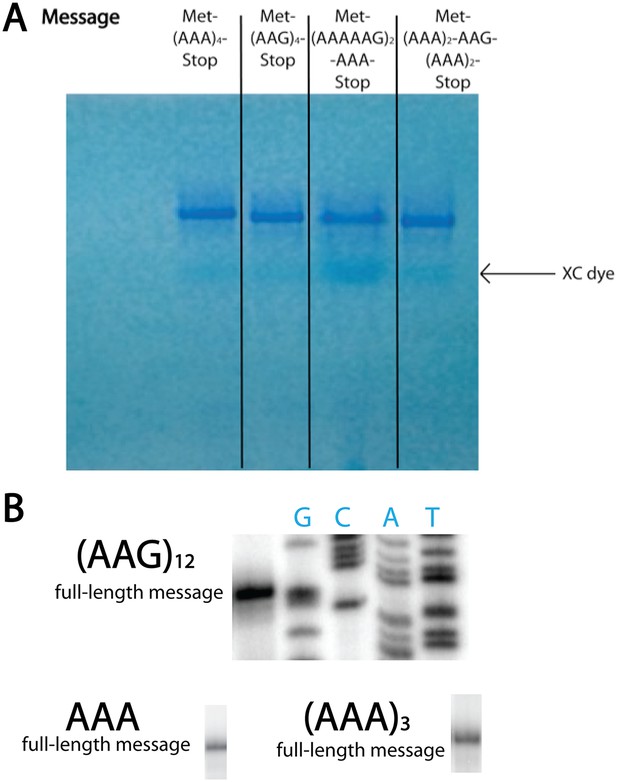
T7 transcribed messages visualized on 15% denaturing PAGE gel.
(A) In vitro transcribed mRNAs used in our in vitro studies run as distinct, single bands on high-resolution denaturing PAGE gel. The RNA is visualized with methylene blue stain. (B) The mRNAs encoding consecutive AAA codons result in discrete length toeprint signatures, yielding specific bands corresponding to the full-length message on our toeprints. We also performed RACE experiments on in vitro T7 transcribed mCherry reporter mRNAs containing A18-36 sequences and found that with high frequency, our RNAs contained the expected number of As. Importantly, in both the cell-free system and in vivo, T7 RNA polymerase is responsible for transcribing the mRNAs relevant to the output. Together, these data provide strong evidence that the mRNAs utilized throughout this study are accurately transcribed by T7 RNA polymerase.
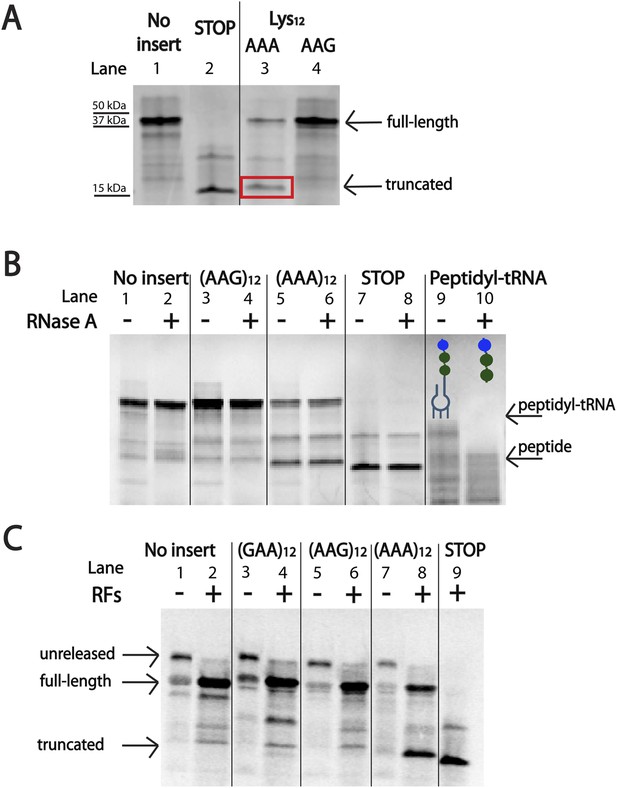
Ribosomes ‘slide’ into new frame on poly(A)-containing messages in the PURE in vitro translation system.
(A) Expression of mCherry reporters (Figure 1A) in the E. coli PURE cell-free translation system (NEB). The truncated band generated from the (AAA)12 reporter is boxed in red. The expected sizes of the full-length, STOP protein and truncated reporter are 42 kDa, 15 kDa, and 17 kDa, respectively. (B) Expression of mCherry reporters in the PURE system and subsequent treatment of peptide products with RNase A. Only the positive control (with a truncated mRNA species) yielded a peptidyl-tRNA product that shifted in mobility upon RNase A treatment. (C) Expression of mCherry reporters (Figure 1A) in the PURE in vitro translation system in the presence and absence of RFs (RFs = RF1, RF2, and RF3).
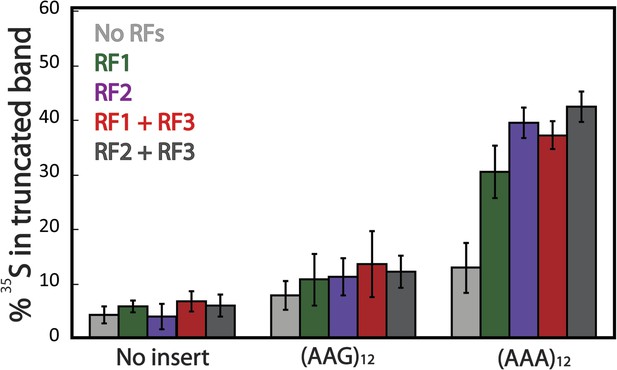
Truncated product release is independent of RF3 in the PURExpress cell-free translation system.
mCherry reporters (Figure 1A: no insert, AAG12, AAA12) were expressed in the PURExpress cell-free translation system lacking release factors (RFs) (light gray). RFs were added back to the reactions individually (RF1 in green, RF2 in purple), and in combination (RF1/3 in red and Rf2/3 dark gray). The plot displays the fraction of protein in the truncated band (100% × (radioactivity in truncated band)/(radioactivity in truncated + full-length bands)).
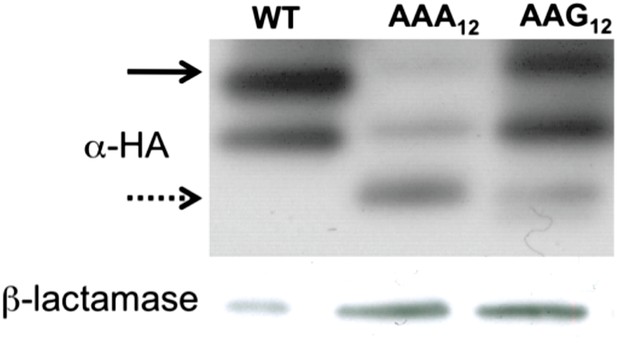
Western blot (α-HA) of mCherry reporters (Figure 1A) expressed in E. coli.
The full-length peptide product is noted with the solid arrow, and the truncated band is highlighted with the dotted arrow. WT = no insert.
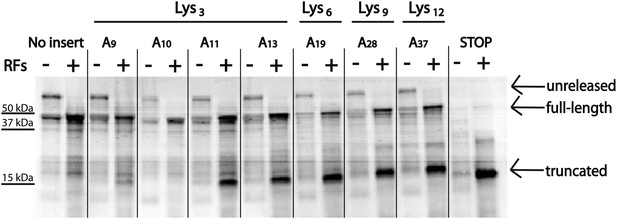
Position and length of poly(A) stretch contributes to ribosome ‘sliding’ in the PURE in vitro translation system.
Expression of mCherry reporters containing poly(A) inserts of various lengths in the presence (+) and absence (−) of RFs.
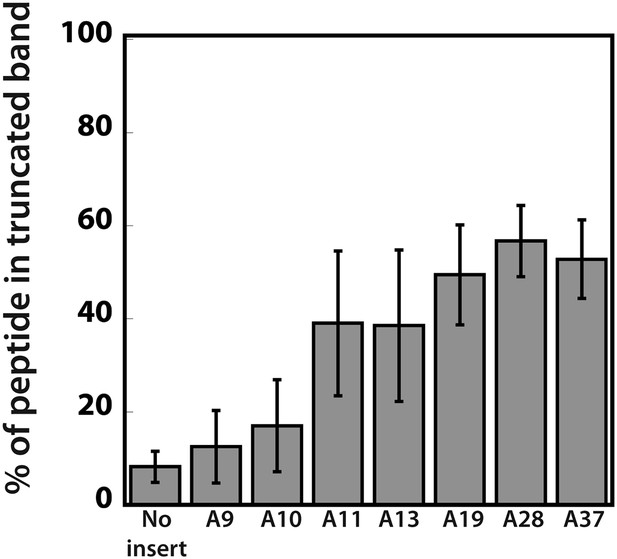
Quantification of the efficiency of ribosome sliding on mCherry reporters expressed in the PURExpress system.
mCherry reporters (Figure 1A: no insert, and various A stretches) were expressed in the PURExpress cell-free translation system (Figure 5). The plot reports the percent of truncated peptide product expressed relative to total peptide product for each reporter (100% × (radioactivity in truncated band)/(radioactivity in truncated + full-length bands)).
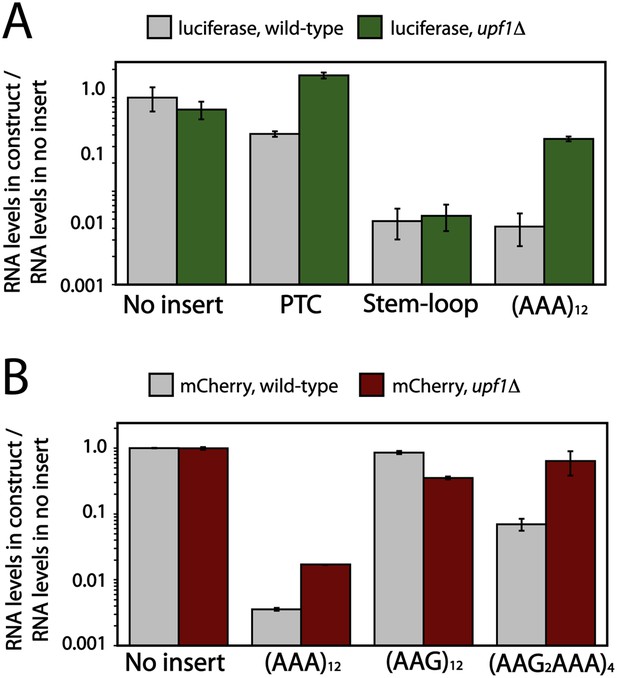
Deletion of Upf1p results in recovery of mRNA levels for poly(A) reporters in yeast.
Luciferase (A) and mCherry (B) reporters (Figure 1A) were expressed in wild-type and upf1Δ S. cerevisiae, and the levels of reporter RNA were quantified by qRT-PCR. Various insertions including 12 lysines ((AAA)12, (AAG)12, (AAG2AAA)4), stem-loop, or premature termination codon (PTC) in the coding sequence are specified on the x-axes.
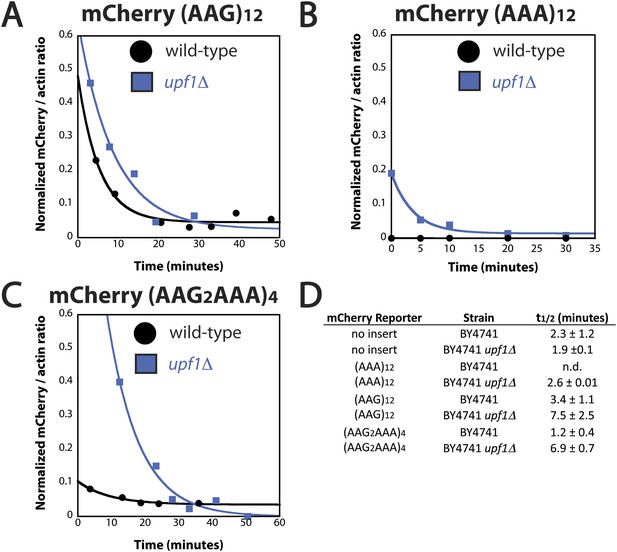
mRNA half-life of reporter containing iterated AAA codons is Upf1 dependent.
Representative experiments measuring the amount of mCherry reporter mRNA in wild-type BY4741 (black) and upf1Δ (blue) cells as a function of time following transcriptional shut-off for reporters containing (A) (AAG)12, (B) (AAA)12, and (C) (AAG2AAA)4 inserts. (D) The measured half-lives for decay of mCherry reporter mRNA in wild-type (BY4741) and upf1Δ cells.
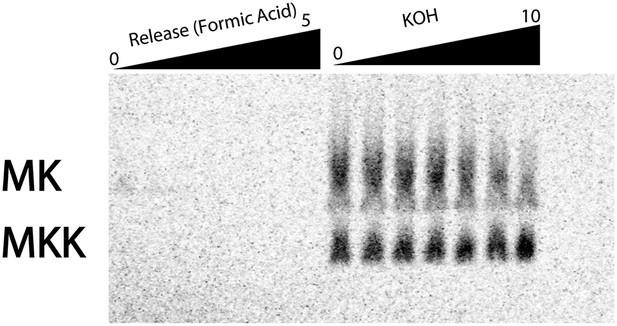
eRF1:3 does not prematurely terminate translation on coding sequences in poly(lysine) messages.
MKA5-STOP message was translated with Lys-tRNALys present by S. cerevisiae ribosomes in a previously described yeast in vitro reconstituted system (Shoemaker, et al., 2010). The reaction was allowed to proceed for 10 min; aliquots of the reaction were quenched at various time points with KOH to hydrolyze the peptidyl-tRNA bond and allow for the visualization of discrete peptide products (right panel). After 10 min, eukaryotic release factors eRf1:eRF3 were added; time points quenched with formic acid; these lanes allow for visualization of peptides released from peptidyl-tRNA (shown in the left panel). Normally, eRF1:eRF3 should only catalyze the release of peptide products from ribosomes on stop codons. The release reaction was allowed to proceed for 5 min (left panel).
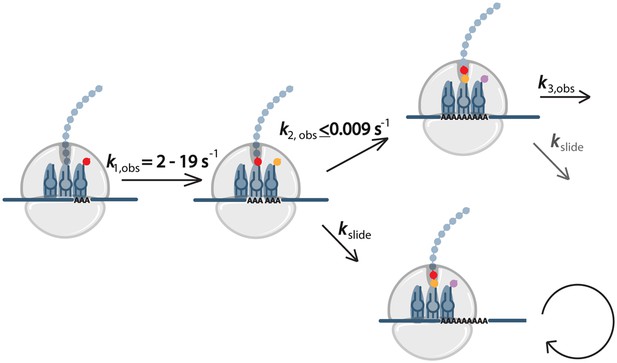
Model for events during ribosome sliding.
In this model translation is paused following the addition of the first lysine. The ribosome can than either slide or perform another round of peptide bond formation. If an AAA codon is positioned in the A site after sliding, the next step will also be slow, while if sliding results in a non-lysine codon in the A site, recovery from slow elongation may occur.
Tables
Bioinformatic analyses of poly(lysine) sequences
| Organism | Sequence | Occurances | Fraction observed | Fraction expected | Enrichment |
|---|---|---|---|---|---|
| E. coli | AAG-AAG | 244 | 0.08 | 0.08 | 1.01 |
| AAG-AAA | 902 | 0.29 | 0.20 | 1.45 | |
| AAA-AAG | 544 | 0.18 | 0.20 | 0.87 | |
| AAA-AAA | 1416 | 0.46 | 0.52 | 0.88 | |
| AAG-AAG-AAG | 9 | 0.07 | 0.02 | 3.37 | |
| AAG-AAG-AAA | 20 | 0.16 | 0.06 | 2.89 | |
| AAG-AAA-AAG | 21 | 0.17 | 0.06 | 3.03 | |
| AAA-AAG-AAG | 4 | 0.03 | 0.06 | 0.58 | |
| AAG-AAA-AAA | 36 | 0.29 | 0.14 | 2.00 | |
| AAA-AAG-AAA | 29 | 0.23 | 0.14 | 1.61 | |
| AAA-AAA-AAG | 4 | 0.03 | 0.14 | 0.22 | |
| AAA-AAA-AAA | 1 | 0.01 | 0.38 | 0.02 | |
| AAG-AAG-AAA-AAG | 1 | 0.25 | 0.02 | 16.07 | |
| AAG-AAG-AAA-AAA | 1 | 0.25 | 0.04 | 6.20 | |
| AAA-AAG-AAA-AAA | 2 | 0.50 | 0.10 | 4.78 | |
| S. cerevisiae | AAG-AAG | 3845 | 0.21 | 0.14 | 1.45 |
| AAG-AAA | 5183 | 0.28 | 0.24 | 1.20 | |
| AAA-AAG | 4505 | 0.24 | 0.24 | 1.04 | |
| AAA-AAA | 4858 | 0.26 | 0.39 | 0.69 | |
| AAG-AAG-AAG | 261 | 0.16 | 0.05 | 2.87 | |
| AAG-AAG-AAA | 234 | 0.14 | 0.09 | 1.57 | |
| AAG-AAA-AAG | 224 | 0.13 | 0.09 | 1.51 | |
| AAA-AAG-AAG | 189 | 0.11 | 0.09 | 1.27 | |
| AAG-AAA-AAA | 211 | 0.13 | 0.15 | 0.87 | |
| AAA-AAG-AAA | 261 | 0.16 | 0.15 | 1.07 | |
| AAA-AAA-AAG | 117 | 0.07 | 0.15 | 0.48 | |
| AAA-AAA-AAA | 171 | 0.10 | 0.24 | 0.43 | |
| AAG-AAG-AAG-AAG | 24 | 0.10 | 0.02 | 4.88 | |
| AAA-AAG-AAG-AAG | 28 | 0.12 | 0.03 | 3.48 | |
| AAG-AAA-AAG-AAG | 23 | 0.10 | 0.03 | 2.86 | |
| AAG-AAG-AAA-AAG | 19 | 0.08 | 0.03 | 2.36 | |
| AAG-AAG-AAG-AAA | 27 | 0.11 | 0.03 | 3.35 | |
| AAG-AAG-AAA-AAA | 13 | 0.05 | 0.06 | 0.99 | |
| AAG-AAA-AAG-AAA | 19 | 0.08 | 0.06 | 1.44 | |
| AAA-AAG-AAG-AAA | 11 | 0.05 | 0.06 | 0.83 | |
| AAA-AAG-AAA-AAG | 17 | 0.07 | 0.06 | 1.29 | |
| AAG-AAA-AAA-AAG | 5 | 0.02 | 0.06 | 0.38 | |
| AAA-AAA-AAG-AAG | 9 | 0.04 | 0.06 | 0.68 | |
| AAG-AAA-AAA-AAA | 9 | 0.04 | 0.09 | 0.42 | |
| AAA-AAG-AAA-AAA | 14 | 0.06 | 0.09 | 0.65 | |
| AAA-AAA-AAG-AAA | 6 | 0.03 | 0.09 | 0.28 | |
| AAA-AAA-AAA-AAG | 5 | 0.02 | 0.09 | 0.23 | |
| AAA-AAA-AAA-AAA | 9 | 0.04 | 0.15 | 0.25 |
-
The prevalence precise sequences encoding 2–3 consecutive lysine residues in E. coli and S. cerevisiae are displayed. The raw number of ‘occurrences’ are listed for each sequence. The enrichment values listed reflect the fraction observed/fraction expected.
Additional files
-
Supplementary file 1
Primary sequence of mCherry with out of frame stop-codons highlighted. The nucleotide sequence of the Thrdx-HA-mCherry reporters (Figure 1A) with all out of frame stop codons after the insertion site highlighted in yellow.
- https://doi.org/10.7554/eLife.05534.021





