Synaptojanin cooperates in vivo with endophilin through an unexpected mechanism
Figures
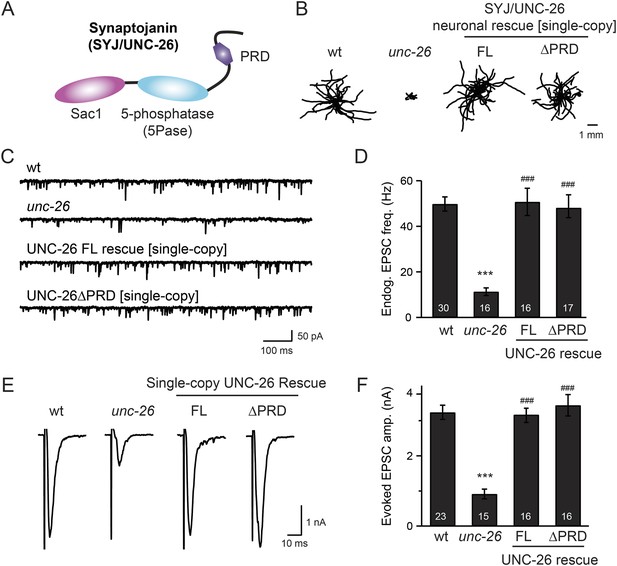
Synaptojanin UNC-26 lacking the PRD domain fully supports locomotion, endogenous activity, and evoked synaptic currents.
(A) Domain structure of synaptojanin UNC-26. Synaptojanin contains three functional modules: a Sac1 phosphatase domain, a 5-phosphatase domain (5Pase), and a proline-rich domain (PRD). Single-copy transgenes encoding GFP-tagged UNC-26 full-length (FL; residues 1–1113) and ∆PRD (residues 1–986) were introduced into synaptojanin unc-26(s1710) mutant worms. The pan-neuronal promoter Prab-3 was used to drive transgene expression. (B) C. elegans locomotion is restored by neuronal expression of full-length synaptojanin (UNC-26FL) or synaptojanin lacking the PRD domain (UNC-26∆PRD). Representative trajectories (20 animals) of 30 s locomotion are shown for each genotype. The starting points for each trajectory are aligned for clarity. (C–F) Electrophysiological recordings show that GFP-tagged synaptojanin UNC-26∆PRD is fully functional at synapses. Representative traces and summary data for endogenous EPSC rates (C–D) and for evoked EPSC amplitude (E–F) are shown for the indicated genotypes. The number of worms analyzed for each genotype is indicated in the bar graphs. ***, p < 0.0001 when compared to wild-type (wt) controls. ###, p < 0.0001 when compared to unc-26 mutants. Error bars represent standard error of the mean (SEM).

Mouse synaptojanin ∆PRD is functional in C. elegans neurons.
Truncated version of mouse synaptojanin 1 (1–1045) that lacks the PRD domain was expressed in C. elegans nervous system under control of the pan-neuronal promoter Psnb-1. Electrophysiological recordings at NMJs were carried out using wt (N2), unc-26(s1710), and transgenic worms carrying mSYJ∆PRD. Summary data for locomotion rates (A), representative traces and summary data for endogenous EPSC rates (B–C), and representative traces and summary data for evoked EPSC amplitude (D–E) are shown for the indicated genotypes. ###, p < 0.0001 when compared to unc-26 mutants. Error bars indicate SEM. The number of worms analyzed for each genotype is indicated in the bar graphs.
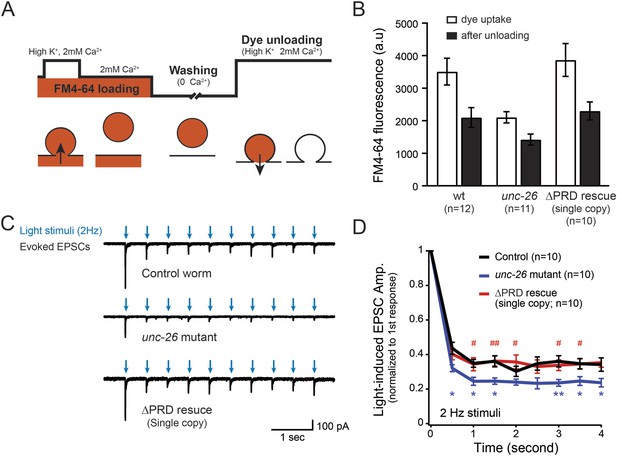
Synaptojanin UNC-26∆PRD recovers the recycling vesicle pool and sustains synaptic transmission upon repetitive stimuli.
(A) A schematic diagram is shown to illustrate the FM4-64 loading and unloading procedure. Experimental details are discussed in the ‘Materials and methods’ section. (B) FM4-64 loading and unloading at the head ganglion were compared for wt (n = 12), unc-26 mutant (n = 11), and rescued worms with a single-copy transgene encoding GFP::UNC-26∆PRD (n = 10). The expression of GFP::UNC-26∆PRD significantly rescued both dye uptake and unloading (p < 0.01 and p < 0.01, respectively; compared to unc-26 mutants). (C) Acetylcholine currents were evoked by 2-Hz light pulses in worms carrying Punc-17::ChR2::mCherry. Representative traces of light-evoked EPSCs during repeated stimulation are shown for the indicated genotypes. (D) Mean values of currents normalized relative to the first EPSC were significantly reduced in unc-26 mutant. The expression of GFP::UNC-26∆PRD in unc-26 mutants restores the amplitude of subsequent currents, suggesting that the UNC-26∆PRD is functional to support synaptic transmission upon repeated stimuli. The number of worms analyzed for each genotype is indicated in the graph. *, p < 0.05 and **, p < 0.01 when compared to wt controls. #, p < 0.05 and ## p < 0.01 when compared to unc-26 mutants. Error bars indicate SEM.
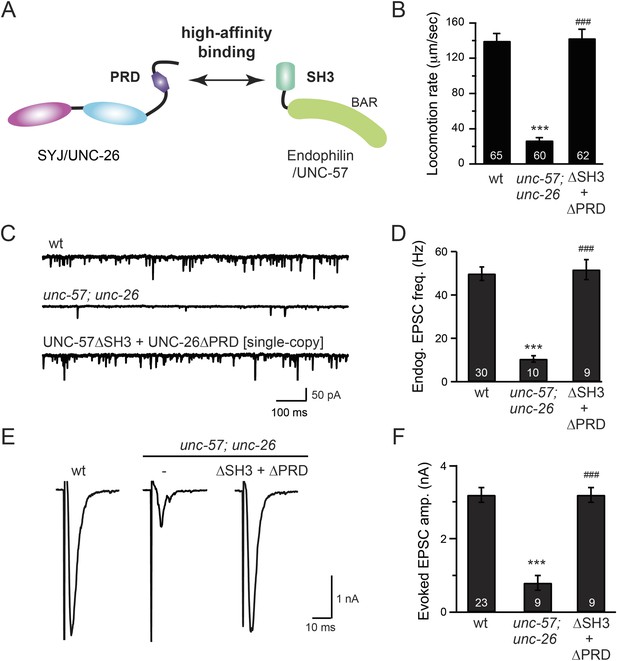
The SH3-PRD interaction is dispensable for synaptic activity.
(A) A schematic drawing showing interactions between synaptojanin (UNC-26) PRD and endophilin (UNC-57) SH3. Single-copy transgenes encoding UNC-26∆PRD::GFP and UNC-57∆SH3::mCherry were co-expressed in unc-57; unc-26 double mutants. Pan-neuronal promoters Prab-3 and Psnb-1 were used to drive expression of UNC-26∆PRD::GFP and UNC-57∆SH3::mCherry, respectively. Summary data for locomotion rate are shown in (B). Representative traces and summary data for endogenous EPSC rates (C–D) and for evoked EPSC amplitude (E–F) are shown for the indicated genotypes. The number of worms analyzed for each genotype is indicated in the bar graphs. ***, p < 0.0001 when compared to wt controls. ###, p < 0.0001 when compared to unc-57; unc-26 double mutants. Error bars indicate SEM.
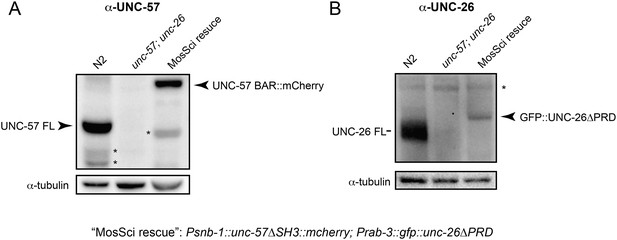
UNC-57 and UNC-26 are not overexpressed in transgenic animals.
Monoclonal antibodies against UNC-57BAR and UNC-26 5-phosphatase were developed as described in the ‘Materials and methods’. (A) Immunoblots for UNC-57 detection. 35 µg of total proteins, extracted from worms of indicated genotypes, were loaded on SDS-PAGE gels. (B) Immunoblots for UNC-26 detection. 100 µg of total proteins were analyzed. Tubulin was used as loading controls and was detected by a monoclonal antibody. Immunoreactive bands were visualized using enhanced chemiluminescence and were quantified using a Bio-Rad ChemiDoc MP imaging system. *, proteolytic products or nonspecific bands.
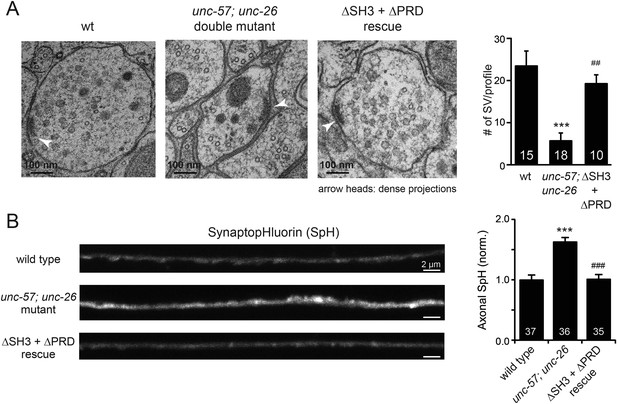
UNC-26∆PRD and UNC-57∆SH3 restore the number of SVs and recover synaptopHluorin retrieval in unc-57; unc-26 double mutants.
(A) Electron microscopy images of neuromuscular junctions were collected from the ventral nerve cords of adult hermaphrodites. Synaptic profiles of 15 synapses of the wt, 18 synapses of the unc-57; unc-26 double mutants, and 10 synapses of the single-copy transgenic UNC-26∆PRD::GFP; UNC-57∆SH3::mCherry animals were analyzed. Arrowheads indicate dense projections. Synaptic vesicle (SV) number was counted in a blind manner. ***, p < 0.0001 when compared to wt controls. ###, p < 0.0001 and ##, p < 0.001 when compared to unc-57; unc-26 double mutants. Scale bar: 100 nm. Error bars indicate SEM. (B) Representative images (left) and summary data (right) for axonal synaptopHluorin (SpH) fluorescence in the dorsal nerve cord are shown for the indicated genotypes. Rescue experiments are done using extrachromosomal arrays carrying Psnb-1::unc-26∆PRD and Prab-3::unc-57∆SH3 (without any fluorescent tags). The number of worms analyzed for each genotype is indicated. ***, p < 0.0001 compared to wt controls. ###, p < 0.0001 when compared to unc-57; unc-26 mutants. Scale bar: 2 µm. Error bars indicate SEM.
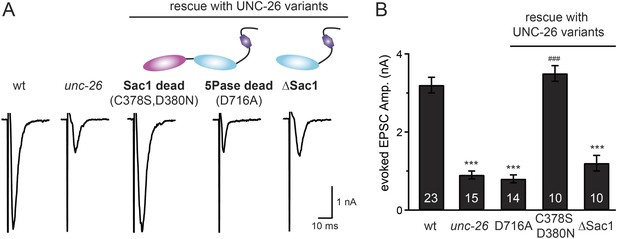
Synaptojanin phosphatase domains have distinct functions.
The 5-Pase domain hydrolyzes phosphoinositides, while Sac1 plays an essential but non-catalytic role at synapses. Evoked EPSCs from wt, unc-26(s1710) mutant, and indicated transgenic strains were compared. Transgenes were GFP-tagged UNC-26 variants, including Sac1 dead (C378S,D380N; full-length UNC-26), 5-Pase dead (D716A, full-length UNC-26), and ∆Sac1 (residues 494–1113). Transgenes were driven by Prab-3. Representative traces (A) and summary data (B) for evoked EPSC amplitudes are shown for the indicated genotypes. ***, p < 0.0001 when compared to wt controls. ###, p < 0.0001 when compared to unc-26 mutants. The number of worms analyzed for each genotype is indicated in the bar graphs. Error bars indicate SEM.
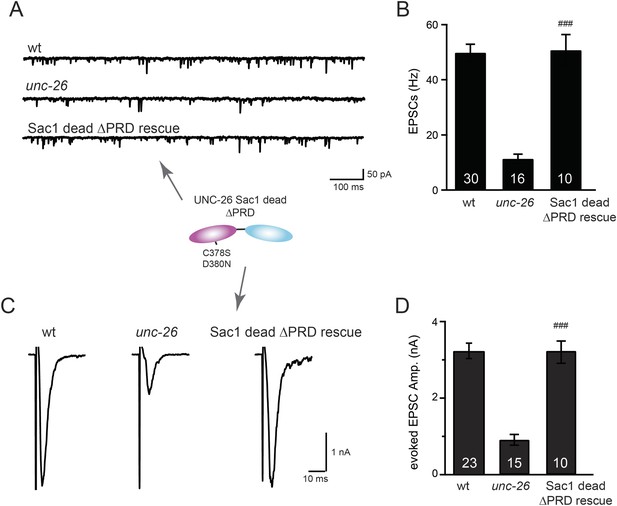
Sac1-inactivated synaptojanin supports synaptic transmission in a PRD independent manner.
Two mutations (C378S and D380N) that inactivate the Sac1 lipid phosphatase activity were introduced into the Sac1 domain of UNC-26∆PRD (residues 1–986). The mutant UNC-26∆PRD (C378S, D380N) was expressed in neurons using Prab-3. Representative traces and summary data for endogenous EPSC rates (A–B) and for evoked EPSC amplitude (C–D) are shown for the indicated genotypes. ###, p < 0.0001 when compared to unc-26 mutants. The number of worms analyzed for each genotype is indicated in the bar graphs. Error bars indicate SEM.
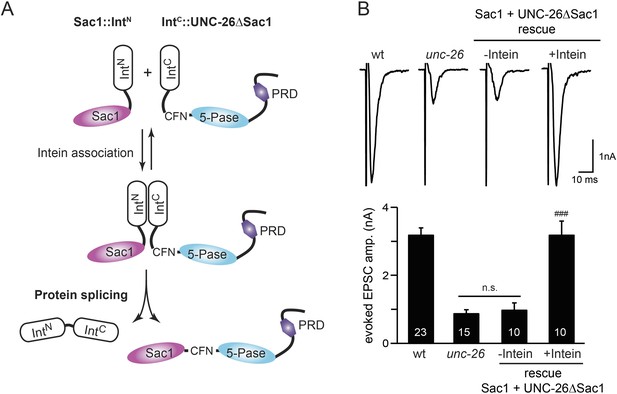
Sac1 must be physically linked to UNC-26 5-phosphatase to support synaptic transmission.
Split-intein mediated ligation was used to post-translationally reconnect Sac1 to the remainder of the UNC-26 synaptojanin protein (A). The Sac1 domain (1–493) was linked to the N-terminal half of NpuDnaE to generate Sac1::IntN. The C-terminal half of NpuDnaE was fused with the N-terminus of the UNC-26∆Sac1 fragment. Three extra residues (CFN) remain in the ligated product. Representative traces and summary data of evoked EPSCs are shown in (B). Co-expression of Sac1::IntN and IntC::UNC-26∆Sac1 significantly rescued the synaptic defects in unc-26 mutant worms. ###, p < 0.0001 when compared to unc-26 mutants. The number of worms analyzed for each genotype is indicated in the bar graphs. Error bars indicate SEM.
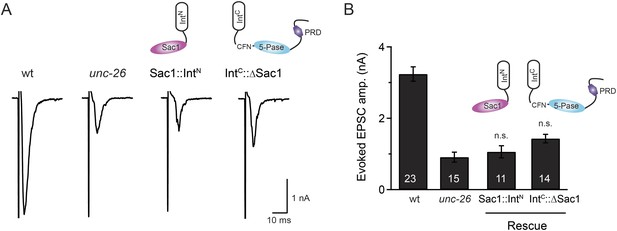
Transgenic worms that only express either Sac1::IntN or IntC::UNC-26∆Sac1 did not show functional improvements.
Representative traces (A) and summary data (B) for evoked EPSC amplitude are shown for the indicated genotypes. Transgenes encoding Sac1::IntN and IntC::UNC-26∆Sac1 were designed as described in the Figure 4. ‘n.s.’ indicates p > 0.05 when compared to unc-26 mutants. The number of worms analyzed for each genotype is indicated in the bar graphs. Error bars indicate SEM.
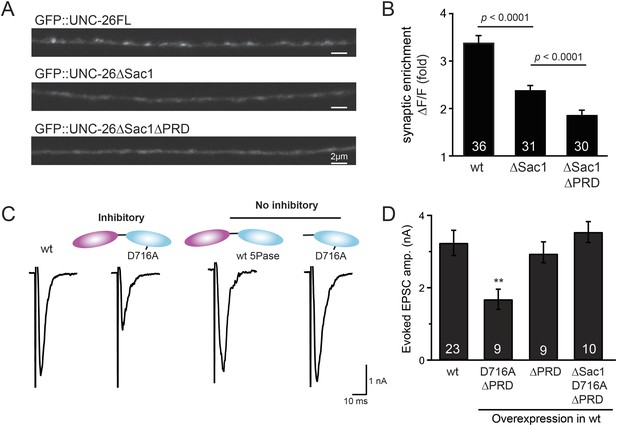
Sac1 is a synaptic targeting domain.
(A–B) Removal of the Sac1 domain of synaptojanin perturbs synaptic targeting of synaptojanin. Representative images (A) showing various versions of GFP::UNC-26 distribution in the dorsal nerve cord. Synaptic enrichment of GFP::UNC-26 was quantified using ∆F/F = (Fpeak − Faxon)/Faxon and was compared for the indicated genotypes (B). Scale bar: 2 µm. (C–D) Sac1 is required for dominant negative inhibition. The D716A mutation that blocks 5-phosphatase activity was introduced into UNC-26∆PRD and UNC-26∆Sac1∆PRD. These UNC-26 variants were expressed in nervous system of wt worms. The stimulus-evoked EPSC amplitudes were significantly reduced in worms carrying UNC-26∆PRD (D716A) mutant proteins. By contrast, animals expressing either UNC-26∆PRD (with a functional 5Pase) or UNC-26∆Sac1∆PRD (D716A) mutant proteins showed normal levels of synaptic activity. The number of worms analyzed for each genotype is indicated in the bar graphs. **, p < 0.001 when compared to wt controls. Error bars represent SEM.
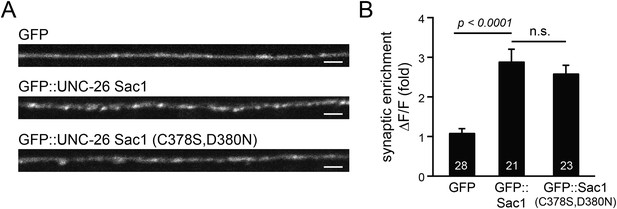
GFP-tagged Sac1 domains localize to synapses.
(A) Representative images showing GFP, GFP::UNC-26Sac1, and GFP::UNC-26Sac1(C378S,D380N) distribution in the dorsal nerve cord axons. Synaptic enrichment of GFP was quantified using ∆F/F = (Fpeak − Faxon)/Faxon. Scale bar: 2 µm. Summary data are shown in (B). The number of worms analyzed for each genotype is indicated in the bar graphs. Error bars indicate SEM.
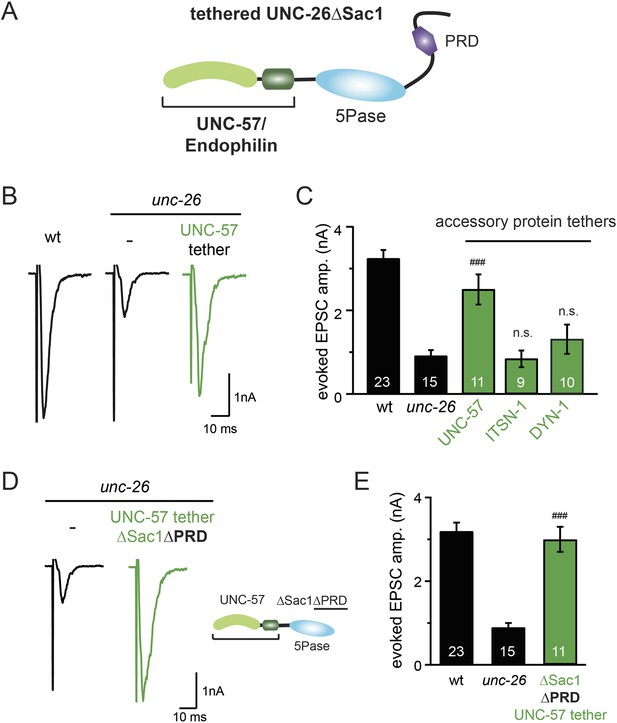
Endophilin functionally substitutes for the Sac1 domain.
(A) A schematic drawing showing the chimeric UNC-57 endophilin::UNC-26∆Sac1 protein. Other endocytic accessory proteins including DYN-1 dynamin and ITSN-1 intersectin were tethered to UNC-26∆Sac1 using an identical strategy. Transgenes were expressed in all neurons using Prab-3. (B–C) Chimeric UNC-57 endophilin::UNC-26∆Sac1 proteins restore evoked EPSCs in unc-26 mutant worms. Other tethers failed to rescue synaptojanin defects. Electrophysiological data in Figure 8B–C and Figure 8—figure supplement 1 were collected blindly. (D–E) The PRD domain is not required for the endophilin tether to bypass the Sac1 requirement of synaptojanin. The number of worms analyzed for each genotype is indicated in the bar graphs. ###, p < 0.0001 when compared to unc-26 mutants. ‘n.s.’ indicates p > 0.05 when compared to unc-26 mutants. Error bars indicate SEM.
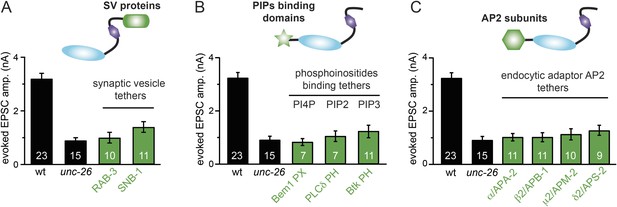
Targeting UNC-26∆Sac1 to synaptic vesicles, phosphoinositides, and endocytic adaptor protein AP2 does not recover synaptojanin function.
Summary data for evoked EPSC amplitudes are shown for the indicated genotypes in (A–C). Non-enzymatic proteins that are involved in SV cycle and phosphoinositide recognition were tethered to the N-terminus of UNC-26∆Sac1, except that SNB-1 and RAB-3 were tethered to the C-terminus of UNC-26∆Sac1. Protein tethers used for these analyses were SV proteins SNB-1/synaptobrevin and RAB-3; phosphatidylinositol lipid-binding domains (PI4P binding Bem1PX domain, PI(4,5)P2 binding PLCδ PH domain, and PI(3,4,5)P3 biding Btk PH domain); and endocytic adaptor AP-2 subunits (APA-2/AP2α, APB-1/AP2β2, APM-2/AP2μ2, and APS-2/AP2δ2). The number of worms analyzed for each genotype is indicated in the bar graphs.
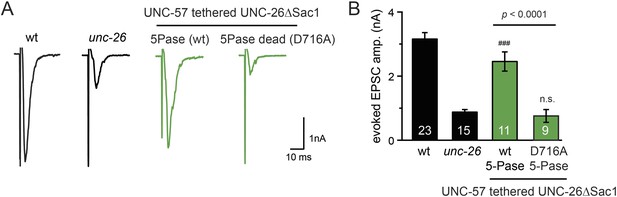
The endophilin tether does not bypass the requirement for a functional synaptojanin 5-phosphatase domain.
The 5-phosphatase activity is required for the UNC-57::UNC-26∆Sac1 chimeric protein. All chimeric proteins were expressed under the control of Prab-3. Representative traces (A) and summary data (B) for evoked EPSC amplitude are shown for the indicated genotypes. The number of worms analyzed for each genotype is indicated in the bar graphs. ###, p < 0.0001, when compared to unc-26 mutants. ‘n.s.’ indicates p > 0.05 when compared to unc-26 mutants. Error bars indicate SEM.
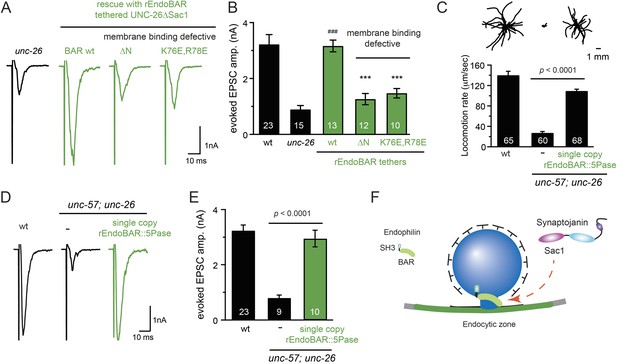
Endophilin BAR domain and its membrane interactions are required for bypassing Sac1.
(A–B) Endophilin BAR is sufficient to bypass the Sac1 requirement. UNC-26∆Sac1 was tethered to worm and rat endophilinA1 BAR (rEndoBAR), respectively. Mutations that disrupt BAR-membrane interactions abolished the rescue activity of the chimeric UNC-26∆Sac1. ***, p < 0.0001 when compared to transgenic unc-26 mutants carrying rEndoBAR wt::UNC-26∆Sac1. (C–E) Expression of a single-copy transgene encoding the rEndoBAR::UNC-26 5Pase chimera significantly restores locomotion and evoked EPSCs in unc-57; unc-26 double mutants. Representative traces (upper) and summary data (lower) for locomotion (C), and evoked EPSCs (D–E) are shown for the indicated genotypes. ###, p < 0.0001 when compared to unc-26 mutants. (F) A schematic diagram showing that the synaptojanin Sac1 domain and the endophilin BAR domain cooperate to promote SV endocytosis.
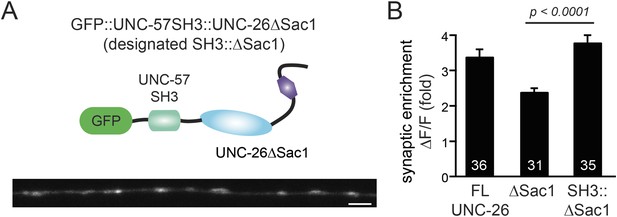
UNC-57 SH3 domain enhances synaptic enrichment of UNC-26∆Sac1.
(A) Distribution of GFP::UNC-57SH3::UNC-26∆Sac1 (domain structure, upper) in the dorsal nerve cord axon is shown (lower). Scale bar: 2 µm. (B) Summary data indicates that UNC-57 SH3 domain enhances the enrichment of UNC-26∆Sac1 at synapses. The number of worms analyzed for each genotype is indicated in the bar graphs. Error bars represent SEM.
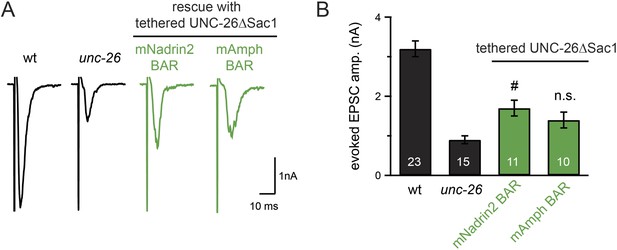
Specificity in BAR proteins for bypassing the Sac1 requirement.
BAR domains of mouse Nadrin2 (residues 1–248) and mouse amphiphysin (residues 1–250) were tethered to the N-terminus of UNC-26∆Sac1. All chimeric proteins were expressed under the control of Prab-3 in unc-26 mutants. Representative traces (A) and summary data (B) for evoked EPSC amplitude are shown for the indicated genotypes. #, p < 0.05 when compared to unc-26 mutants. Error bars represent SEM. n.s. indicates non-significant (p = 0.07, compared with unc-26).

UNC-26 Sac1 does not bind UNC-57 in solution.
GST pull-down assays were performed as described in the ‘Materials and methods’. Briefly, GST::UNC-57 (10 µg) was immobilized on glutathione beads. Maltose-binding protein (MBP)::UNC-26 fragments were incubated with beads for 2 hr. Twenty percent of samples and 7% of total MBP::UNC-26 fragments were subjected to SDS-PAGE and visualized by staining with Coomassie Brilliant Blue G-250. The Sac1 domain did not bind to full-length UNC-57, nor UNC-57 BAR (left panel). The 5-phosphatase domain also did not exhibit UNC-57 binding (middle panel). In contrast, the PRD domain exhibited a significant amount of binding to UNC-57 SH3 (right panel).
Tables
Summary of data from electrophysiological recordings and locomotion analyses
| Evoked EPSC Amp. (nA) | Endogenous EPSC | Locomotion speed (µm/s) | |||
|---|---|---|---|---|---|
| Frequency (Hz) | Amp. (pA) | ||||
| Wild type (N2) | 3.2 ± 0.2 (n = 23) | 49.8 ± 2.5 (n = 30) | 22.4 ± 1.0 | 140 ± 8 (n = 65) | |
| unc-26 (s1710) | – | 0.9 ± 0.1 (n = 15)† | 12.0 ± 1.5 (n = 16)† | 20.7 ± 0.6 | 28 ± 3 (n = 65)† |
| Si[Prab-3::unc-26::gfp] | 3.1 ± 0.2 (n = 16)# | 50.8 ± 5.4 (n = 16)# | 23.2 ± 1.4 | 139 ± 8 (n = 60)# | |
| Si[Prab-3::unc-26∆PRD::gfp] | 3.4 ± 0.3 (n = 16)# | 47.4 ± 5.0 (n = 17)# | 24.8 ± 1.3 | 141 ± 10 (n = 60)# | |
| Ex[Psnb-1::mSYJ1∆PRD] | 3.1 ± 0.3 (n = 7)# | 49.6 ± 6.1 (n = 11)# | 21.5 ± 0.6 | 143 ± 10 (n = 37)# | |
| Ex[Prab-3::gfp::unc-26(C378S,D380N)] | 3.5 ± 0.2 (n = 10)# | 53.1 ± 6.6 (n = 10) | 23.8 ± 1.3 | 135 ± 9 (n = 60)# | |
| Ex[Prab-3::gfp::unc-26∆PRD(C378S,D380N)] | 3.2 ± 0.3 (n = 10)# | 50.7 ± 5.7 (n = 10) | 24.2 ± 0.9 | 130 ± 9 (n = 60)# | |
| Ex[Prab-3::gfp::unc-26(D716A)] | 0.8 ± 0.1 (n = 14) | 10.0 ± 1.8 (N = 15) | 20.9 ± 0.8 | 28 ± 3 (n = 60) | |
| Ex[Prab-3::gfp::unc-26∆Sac1] | 1.2 ± 0.2 (n = 10) | 7.2 ± 1.9 (N = 10) | 21.3 ± 1.2 | 31 ± 3 (n = 60) | |
| Ex[Prab-3::unc-26Sac1 + Prab-3::unc-26∆Sac1] | 1.0 ± 0.2 (n = 10) | 16.1 ± 1.8 (n = 10) | 20.4 ± 0.5 | 32 ± 3 (n = 60) | |
| Ex[Prab-3::unc-26Sac1::IntN + Prab-3::IntC::unc-26∆Sac1] | 3.2 ± 0.4 (n = 10)# | 53.1 ± 7.1 (n = 10)# | 24.8 ± 1.2 | 100 ± 7 (n = 60)# | |
| Ex[Prab-3::unc26∆Sac1::rab-3] | 1.0 ± 0.2 (n = 10) | 13.2 ± 2.3 (n = 10) | 20.4 ± 0.9 | ||
| Ex[Prab-3::unc-26∆Sac1::snb-1] | 1.4 ± 0.2 (n = 11) | 17.9 ± 2.6 (n = 11) | 21.1 ± 0.8 | ||
| Ex[Prab-3::bem1PX::unc-26∆Sac1] | 0.8 ± 0.1 (n = 7) | 6.3 ± 0.6 (n = 7) | 18.8 ± 1.2 | ||
| Ex[Prab-3::plc∂PH::unc-26∆Sac1] | 1.0 ± 0.2 (n = 7) | 8.5 ± 1.8 (n = 7) | 18.7 ± 1.1 | ||
| Ex[Prab-3::btkPH::unc-26∆Sac1] | 1.2 ± 0.2 (n = 11) | 11.4 ± 1.2 (n = 11) | 18.4 ± 0.8 | ||
| Ex[Prab-3::apa-2::unc-26∆Sac1] | 1.1 ± 0.1 (n = 11) | 12.8 ± 1.8 (n = 11) | 19.4 ± 0.8 | ||
| Ex[Prab-3::apb-1::unc-26∆Sac1] | 0.9 ± 0.1 (n = 11) | 13.2 ± 2.5 (n = 11) | 18.2 ± 1.5 | ||
| Ex[Prab-3::apm-2::unc-26∆Sac1] | 1.1 ± 0.1 (n = 10) | 15.3 ± 2.7 (n = 10) | 21.0 ± 0.9 | ||
| Ex[Prab-3::aps-2::unc-26∆Sac1] | 1.3 ± 0.2 (n = 9) | 12.7 ± 1.4 (n = 9) | 20.1 ± 0.9 | ||
| Ex[Prab-3::unc-57::unc-26∆Sac1] | 2.5 ± 0.3 (n = 11)# | 28.0 ± 4.1 (n = 11)§ | 23.0 ± 1.4 | ||
| Ex[Prab-3::dyn-1::unc-26∆Sac1] | 1.3 ± 0.3 (n = 10) | 14.4 ± 2.9 (n = 10) | 19.4 ± 0.9 | ||
| Ex[Prab-3::itsn-1::unc-26∆Sac1] | 0.9 ± 0.1 (n = 9) | 13.9 ± 1.4 (n = 9) | 21.5 ± 1.4 | ||
| Ex[Prab-3::unc-57::unc-26∆Sac1∆PRD] | 3.0 ± 0.3 (n = 11)# | 36.7 ± 5.4 (n = 11)§ | 21.7 ± 1.3 | ||
| Ex[Prab-3::unc-57::unc-26∆Sac1(D716A)] | 0.8 ± 0.2 (n = 9) | 13.0 ± 2.3 (n = 9) | 21.0 ± 0.7 | ||
| Ex[Prab-3::unc-57BAR::unc-26∆Sac1] | 2.9 ± 0.2 (n = 12)# | 23.9 ± 2.5 (n = 12)# | 21.3 ± 0.8 | ||
| Ex[Prab-3::rEndoBAR::unc-26∆Sac1] | 3.1 ± 0.4 (n = 13)# | 28.5 ± 4.6 (n = 13)# | 24.1 ± 1.4 | ||
| Ex[Prab-3::mAmphBAR::unc-26∆Sac1] | 1.4 ± 0.2 (n = 10) | 19.2 ± 2.3 (n = 10) | 20.8 ± 0.8 | ||
| Ex[Prab-3::mNadrin2BAR::unc-26∆Sac1] | 1.7 ± 0.2 (n = 11)‡ | 16.1 ± 3.1 (n = 11) | 23.8 ± 1.9 | ||
| Ex[Prab-3::rEndoBAR∆N::unc-26∆Sac1] | 1.3 ± 0.2 (n = 12) | 9.1 ± 0.9 (n = 12) | 18.9 ± 0.4 | ||
| Ex[Prab-3::rEndoBAR(K76E,K78E)::unc-26∆Sac1] | 1.5 ± 0.2 (n = 10) | 13.5 ± 1.3 (n = 10) | 19.3 ± 0.7 | ||
| N2 | Prab-3::unc-26∆PRD(D716A) overexpression | 1.5 ± 0.3 (n = 9)* | 24.6 ± 3.8 (n = 10)* | 25.7 ± 0.9 | |
| Prab-3::unc-26∆PRD overexpression | 2.9 ± 0.3 (n = 9) | 53.5 ± 4.6 (n = 9) | 25.4 ± 1.3 | ||
| Prab-3::unc-26∆Sac1∆PRD(D716A) overexpression | 3.5 ± 0.3 (n = 10) | 49.0 ± 7.5 (n = 10) | 25.9 ± 1.9 | ||
| unc-57(e406); unc-26(s1710) | – | 0.8 ± 0.2 (n = 9)† | 8.6 ± 0.8 (n = 10)† | 21.9 ± 1.1 | 27 ± 3 (n = 60)† |
| Si[Psnb-1::unc-57∆SH3::mCherry]; Si[Prab-3::unc-26∆PRD::gfp] | 3.2 ± 0.2 (n = 9)¶ | 50.3 ± 4.1 (n = 9)¶ | 23.1 ± 1.0 | 142 ± 9 (n = 62)¶ | |
| Si[Psnb-1::rEndoBAR::unc-26∆Sac1∆PRD] | 3.0 ± 0.3 (n = 10)¶ | 50.9 ± 4.1 (n = 10)¶ | 26.5 ± 1.1 | 109 ± 4 (n = 68)¶ | |
-
*
p < 0.001 when compared with N2.
-
†
p < 0.0001 when compared with N2.
-
‡
p < 0.05 when compared with unc-26 mutant.
-
§
p < 0.001 when compared with unc-26 mutant.
-
#
p < 0.0001 when compared with unc-26 mutant.
-
¶
p < 0.0001 when compared with unc-57; unc-26 double mutants.
-
Si: single-copy transgene (MosSci insertion).
-
Ex: extrachromosomal array.
-
‘Amp.’ indicates amplitude.





