A non-canonical mechanism for Crm1-export cargo complex assembly
Figures
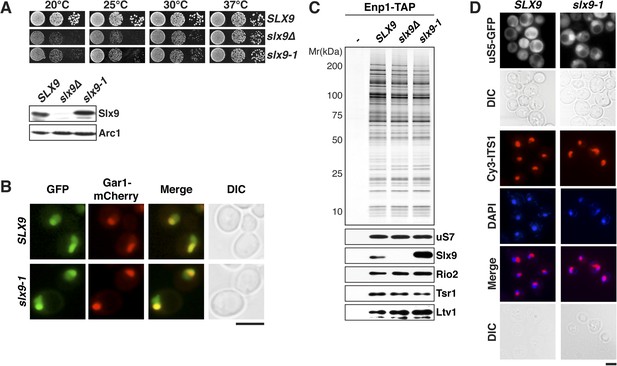
slx9-1 phenocopies the slx9∆ mutation.
(A) The slx9-1 allele does not complement the slow growth of slx9∆ cells. Top: SLX9, slx9∆, and slx9-1 cells were spotted in 10-fold dilutions on SD-plates and grown at the indicated temperatures for 3–6 days. Bottom: Slx9 protein levels from whole cell extracts derived from the indicated strains were determined by Western analysis using antibodies directed against Slx9. Levels of the protein Arc1 served as a loading control. (B) Slx9-1 localizes to the nucleolus/nucleoplasm. Cells expressing Gar1-mCherry and Slx9-GFP or Slx9-1-GFP were grown until mid-log phase. Localization of the indicated fusion proteins was analyzed by fluorescence microscopy. Gar1-mCherry served as a nucleolar marker. Scale bar = 5 µm. (C) Slx9-1 is recruited to the early 40S pre-ribosome. Enp1-TAP was isolated by tandem affinity purification (TAP) from the indicated strains. Calmodulin-eluates were separated on a 4–12% gradient gel and analyzed by either silver staining or Western using the indicated antibodies. The ribosomal protein uS7 served as a loading control. (D) slx9-1 cells are impaired in nuclear export of 40S pre-ribosomes. Top: localization of uS5-GFP was monitored by fluorescence microscopy. Bottom: localization of 20S pre-rRNA was analyzed by FISH using a Cy3-labeled oligonucleotide complementary to the 5′ portion of ITS1 (red). Nuclear and mitochondrial DNA was stained by DAPI (blue). Scale bar = 5 µm.
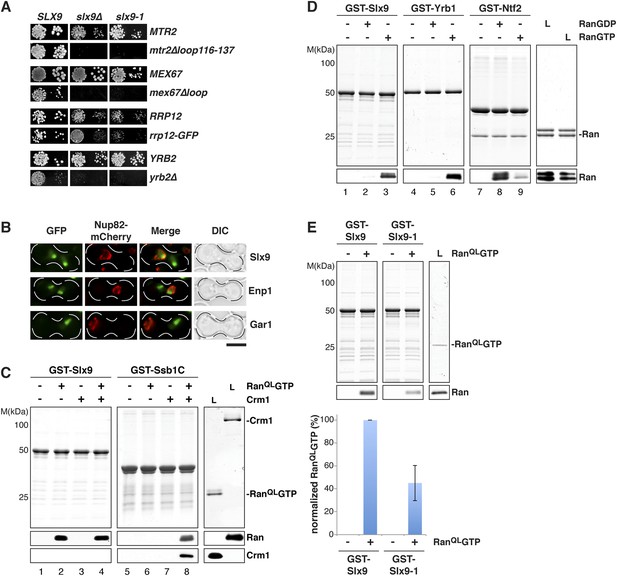
Slx9 is a RanGTP binding protein.
(A) slx9-1 genetically interacts with factors involved in 40S pre-ribosome export. slx9-1 is synthetically lethal with mex67∆loop, mtr2∆loop116-137, or yrb2∆ and strongly synthetically enhanced with rrp12-GFP. Strains containing the indicated WT and mutant alleles were spotted in 10-fold serial dilutions on 5-FOA-SD or SD and grown at 20–30°C for 3–6 days. (B) Slx9 shuttles between the nucleus and the cytoplasm. Cells expressing Enp1-GFP, Gar1-GFP, or Slx9-GFP were mated with kar1-1 cells expressing Nup82-mCherry. The resulting heterokaryons were analyzed by fluorescence microscopy. Scale bar = 5 µm. (C) Slx9 directly binds to RanGTP. GST-Slx9 or GST-Ssb1C was immobilized on GSH-Sepharose before incubating with either buffer alone or buffer containing 2 µM His6-RanQLGTP, 50 nM Crm1-His6 or 2 µM His6-RanQLGTP, and 50 nM Crm1-His6. After washing, bound proteins were eluted in LDS sample buffer, separated by SDS-PAGE and visualized by Coomassie staining or Western blotting using the indicated antibodies. L = input. (D) Slx9 specifically interacts with the GTP-bound form of Ran. GST-Slx9, GST-Yrb1, or GST-Ntf2 was immobilized on GSH-Sepharose and incubated with buffer alone or 2 µM His6-Ran loaded with GDP or GTP. Analysis of the eluted proteins was carried out as described in (C). L = input. (E) Slx9-1 binding to RanGTP is impaired. Top: GST-Slx9 or GST-Slx9-1 immobilized on GSH-Sepharose was incubated with buffer alone or 2 µM His6-RanQLGTP. Analysis of the eluted proteins was carried out as described in (C). L = input. Bottom: bar graph depicts the bound His6-RanQLGTP Western blot signal normalized to GST-Slx9 and GST-Slx9-1 levels, respectively. Four independent experiments were performed and Western blots were quantified by software ImageJ (Version 1.44o). Error bars (S.D.) are indicated.
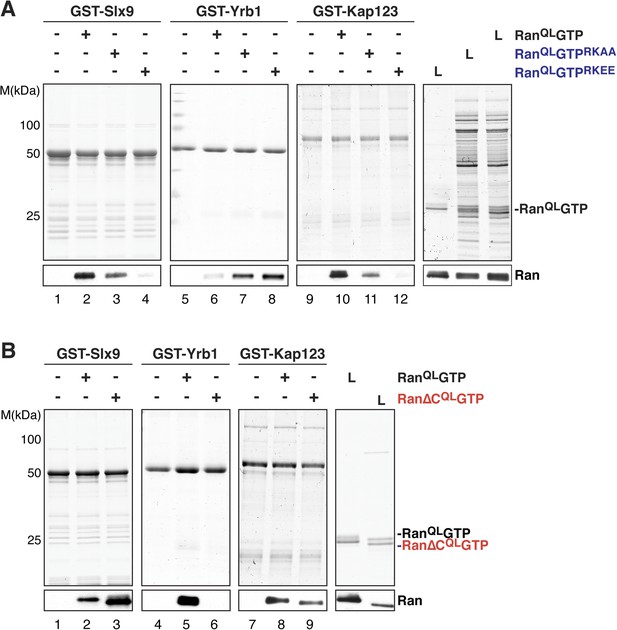
The basic patch and acidic tail of Ran modulates interactions with Slx9.
(A) The basic patch of RanQLGTP contributes to Slx9 binding. GST-Slx9, GST-Yrb1, or GST-Kap123 immobilized on GSH-Sepharose was incubated with buffer alone or 2 µM Ran (His6-RanQLGTP, His6-RanQLGTPRKAA, or His6-RanQLGTPRKEE). After washing, bound proteins were eluted in LDS sample buffer, separated by SDS-PAGE and visualized by Coomassie staining or Western blotting using the indicated antibody. L = input. (B) The acidic tail of RanQLGTP negatively regulates interactions with Slx9. GST-Slx9, GST-Yrb1, or GST-Kap123 immobilized on GSH-Sepharose was incubated with buffer alone or 2 µM Ran (His6-RanQLGTP or His6-Ran∆CQLGTP). Analysis of the eluted proteins was carried out as described in (A). L = input.
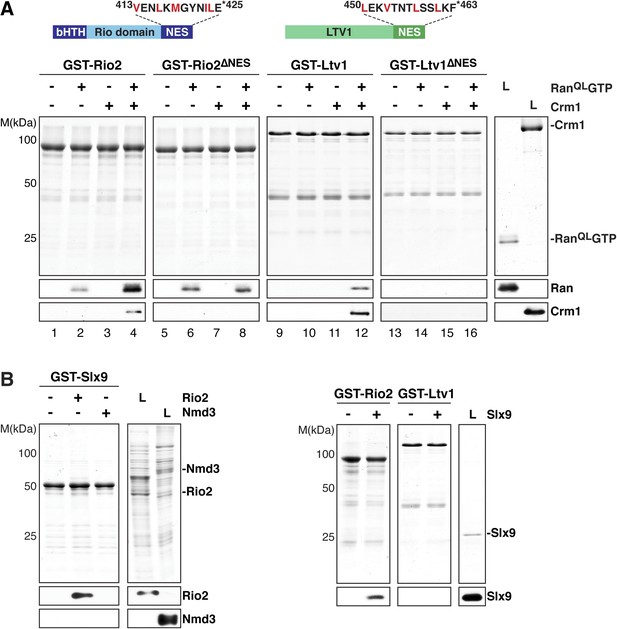
Slx9 directly binds the 40S pre-ribosome nuclear export signal (NES)-containing adaptor Rio2 and RanQLGTP.
(A) Rio2 and Ltv1 export complex formation requires their C-terminal NESs. Top: the positions of the Rio2 and Ltv1 NESs are shown. Hydrophobic residues in these NESs are highlighted in red. Bottom: GST-Rio2, GST-Rio2∆NES, GST-Ltv1, or GST-Ltv1∆NES was immobilized on GSH-Sepharose, and complex formation was analyzed as in Figure 2C. L = input. (B) Slx9 directly interacts with Rio2. Immobilized GST-Rio2 or GST-Ltv1 was incubated with buffer alone or 0.5 µM Slx9. Conversely, immobilized GST-Slx9 was incubated with buffer alone or with lysate containing His6-Nmd3 or His6-Rio2. Analysis of the eluted proteins was carried out as described in Figure 2C. L = input.
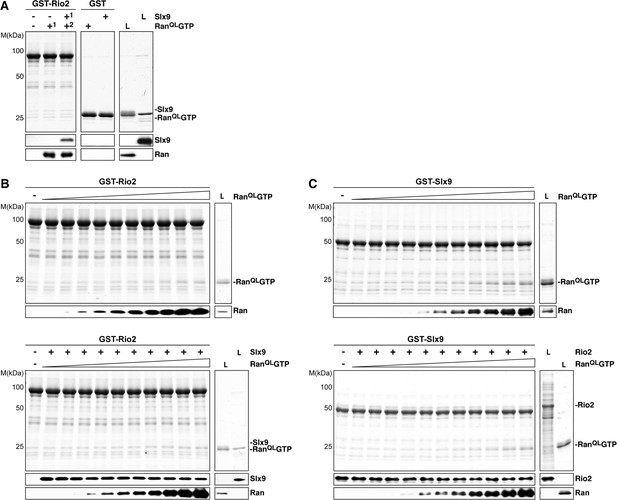
Slx9 binds to Rio2 and RanGTP using distinct binding surfaces.
(A) GST-Rio2 was immobilized on GSH-Sepharose and incubated with buffer, 2 µM His6-RanQLGTP, or 0.5 µM Slx9 (+1). After washing, the GST-Rio2:Slx9 complex was incubated with 2 µM His6-RanQLGTP (+2). Analysis of the eluted proteins was carried out as described in Figure 2C. L = input. (B) RanGTP does not displace Slx9 from a preformed GST-Rio2:Slx9 complex. Top: immobilized GST-Rio2 was incubated with buffer or increasing concentrations of His6-RanQLGTP (62.5 nM–32 µM). Bottom: immobilized GST-Rio2 was incubated with either buffer or 1 µM Slx9. The unbound Slx9 was washed away, and the resulting GST-Rio2:Slx9 complex was incubated with increasing concentrations of His6-RanQLGTP (62.5 nM–32 µM). Analysis of the eluted proteins was carried out as described in Figure 2C. L = input. (C) RanQLGTP does not displace Rio2 from a preformed GST-Slx9:Rio2 complex. Top: immobilized GST-Slx9 was incubated with buffer or increasing concentrations of His6-RanQLGTP (62.5 nM–32 µM). Bottom: immobilized GST-Slx9 was incubated with excess of Rio2. The unbound Rio2 was washed away, and the resulting complex GST-Slx9:Rio2 complex was incubated with increasing concentrations of His6-RanQLGTP (62.5 nM–32 µM). Analysis of the eluted proteins was carried out as described in Figure 2C. L = input.
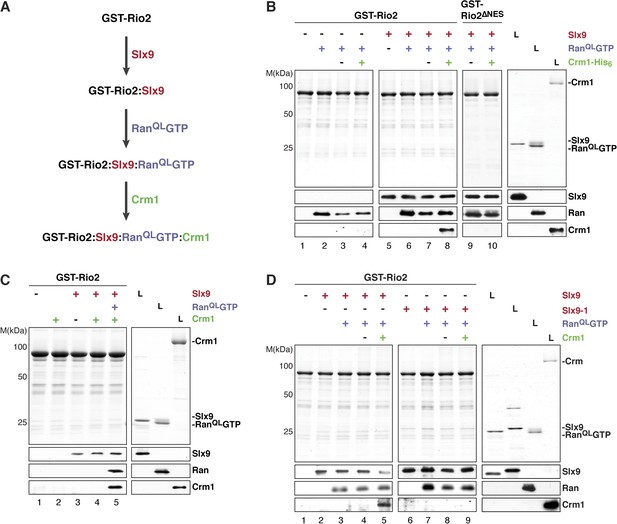
Slx9 promotes stepwise assembly of a Crm1-export complex on the NES of Rio2.
(A) Flow chart depicting the experimental setup to assemble a Rio2:Slx9:RanQLGTP:Crm1 complex. Immobilized GST-Rio2 was sequentially incubated with Slx9 (red), RanQLGTP (purple), and Crm1 (green). Unbound protein was washed away after each incubation step. (B) Crm1 is recruited to the GST-Rio2:Slx9:RanGTP complex in a NES-dependent manner. Immobilized GST-Rio2 or GST-Rio2∆NES was incubated with buffer alone or 0.5 µM Slx9, followed by the stepwise addition of 0.2 µM His6-RanQLGTP and 50 nM Crm1-His6, as depicted in (A). After a final washing step, bound proteins were analyzed as in Figure 2C. L = input. (C) Crm1 is not recruited to the GST-Rio2:Slx9 complex. Immobilized GST-Rio2 was incubated with buffer alone or 0.5 µM Slx9, followed by addition of buffer, 50 nM Crm1-His6, or the stepwise addition of 0.2 µM His6-RanQLGTP and 50 nM Crm1-His6 as depicted in (A). Analysis of the bound proteins was carried out as described in Figure 2C. L = input. (D) Recruitment of Crm1 to a Rio2:Slx9-1:RanQLGTP complex is impaired. Immobilized GST-Rio2 was incubated with buffer alone, 0.5 µM Slx9 or 0.5 µM Slx9-1, followed by the stepwise addition of 0.2 µM His6-RanQLGTP and 50 nM Crm1-His6 as depicted in (A). Analysis of the bound proteins was carried out as described in Figure 2C. L = input.
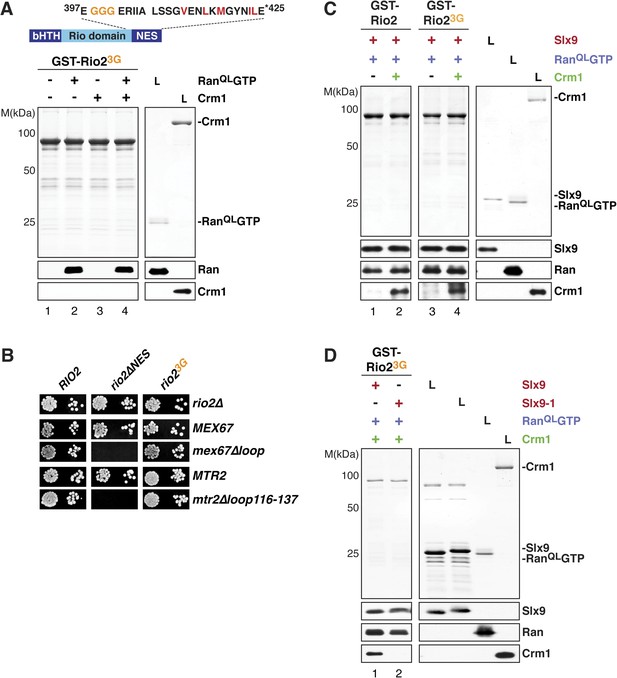
Slx9 provides a scaffold to load Crm1 onto Rio2-NES.
(A) Rio23G does not interact with Crm1 in the presence of RanGTP. Top: schematic depicts the positions of mutations proximal to the NES (399-EEN-401-GGG) in the Rio23G. Hydrophobic amino acids of the NES are red and mutated amino acids are orange. Bottom: GST-Rio23G was immobilized on GSH-Sepharose and binding reactions were carried out and analyzed as in Figure 2C. L = input. (B) rio2∆NES, but not rio23G, is synthetically lethal with mex67∆loop and mtr2∆loop116-137. Strains were spotted in 10-fold serial dilutions on 5-FOA (SD) plates and grown at 30°C for 2–4 days. (C) Slx9 restores Crm1 binding to the Rio23G:Slx9:RanQLGTP complex. GST-Rio2:Slx9:RanQLGTP or GST-Rio23G:Slx9:RanQLGTP was incubated with buffer alone or 50 nM Crm1-His6. Bound proteins were analyzed as in Figure 2C. L = input. (D) Crm1 is impaired in binding a Rio23G:Slx9-1:RanQLGTP complex. Immobilized GST-Rio23G was incubated with buffer alone, 0.5 µM Slx9 or 0.5 µM Slx9-1, followed by the stepwise addition of 0.2 µM His6-RanQLGTP and 50 nM Crm1-His6 as depicted in (A). Analysis of the bound proteins was carried out as described in Figure 2C. L = input.
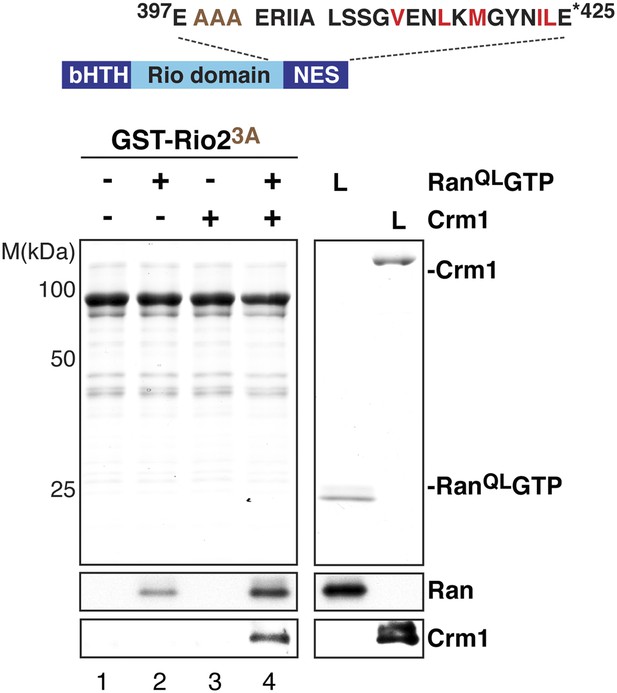
The ‘flexibility’ of the NES region in Rio2 contributes to its interaction with Crm1 in the presence of RanGTP.
Top: schematic of Rio2 highlighting the triple A mutation (399-EEN-401-AAA, brown) proximal to the NES. Hydrophobic amino acids of the NES are red and mutated amino acids are brown. Bottom: immobilized GST-Rio23A was incubated with buffer alone or buffer containing 2 µM His6-RanQLGTP, 50 nM Crm1-His6 or 2 µM His6-RanQLGTP, and 50 nM Crm1-His6. After washing, eluted proteins were separated by SDS-PAGE and visualized by Coomassie staining or Western blotting using indicated antibodies. L = input.

Genetic interactions between Rio2 alleles and RanGTP-binding proteins Slx9 and Yrb2.
(A) slx9∆ and slx9-1 do not genetically interact with rio2∆NES. A RIO2 shuffle slx9∆ strain was transformed with the indicated combinations of empty, WT, or mutant plasmids and spotted in 10-fold dilutions on SD-plates containing 5-FOA and grown at 25°C for 2–4 days. (B) rio2∆NES weakly genetically interacts with yrb2∆. RIO2 shuffle yrb2∆ strain transformed with the indicated combinations of empty, WT or mutant plasmids were spotted in 10-fold dilutions on SD-plates containing 5-FOA and grown at 25°C for 2–4 days.
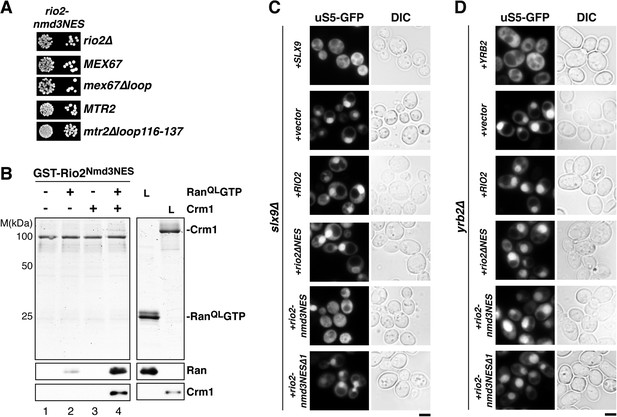
Strong NESs of Nmd3 on Rio2 bypass requirement for Slx9 but not Yrb2 in 40S pre-ribosome export.
(A) rio2-nmd3NES is not synthetic lethal with mex67∆loop or mtr2∆loop116-137. Strains were spotted in 10-fold serial dilutions on 5-FOA (SD) plates and grown at 30°C for 2–4 days (B) The Nmd3-NES (amino acids 440–518) fused to Rio2∆NES bypasses the requirement of the Rio2-NES in export complex formation in vitro. GST-Rio2Nmd3NES was immobilized on GSH-Sepharose and complex formation was carried out and analyzed as in Figure 2C. L = input. (C) rio2-nmd3NES rescues the impaired pre40S export of slx9∆ cells. Localization of uS5-GFP in the indicated strains was monitored by fluorescence microscopy. Scale bar = 5 µm. (D) The rescue of impaired pre40S ribosome export by rio2-nmd3NES is specific for slx9∆. yrb2∆ cells transformed with the indicated plasmids was monitored by fluorescence microscopy for the localization of uS5-GFP. Scale bar = 5 µm.

Comparison of Coomassie staining and Western blot signals of Slx9 and Slx9-1 proteins. 0.5 µM Slx9 or 0.5 µM Slx9-1 were separated on SDS-PAGE (top panel) and either stained with Coomassie or detected by Western analysis with Slx9 antibody (lower panel). L = input.
Additional files
-
Supplementary file 1
Yeast strains used in this study.
- https://doi.org/10.7554/eLife.05745.013
-
Supplementary file 2
Plasmids used in this study.
- https://doi.org/10.7554/eLife.05745.014





