Extensive site-directed mutagenesis reveals interconnected functional units in the alkaline phosphatase active site
Figures
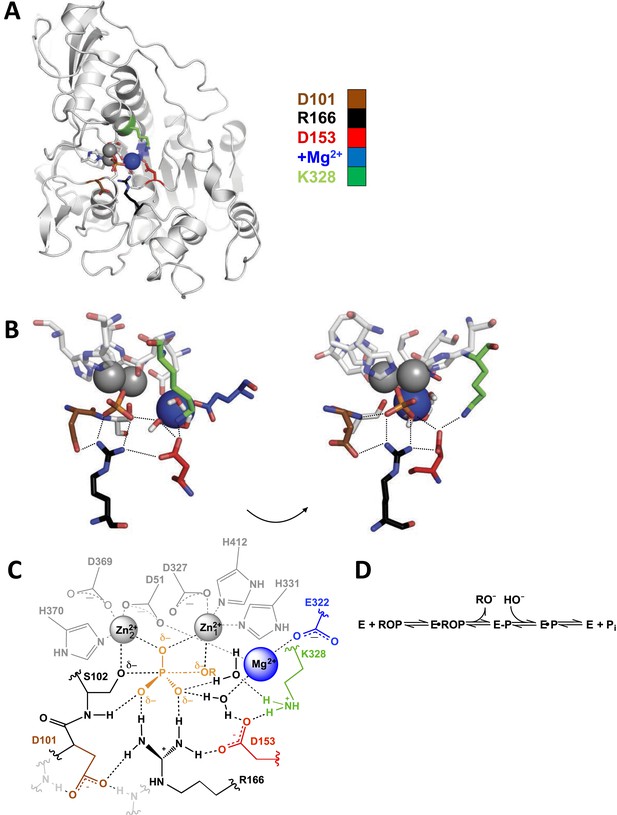
Alkaline phosphatase (AP) structure and active site.
(A) The three-dimensional structure of AP with bound Pi (PDB 3TG0). Active site residues are depicted as follows: D101, brown; R166, black; D153, red; K328, green; E322 and Mg2+ ion, blue. (B) A close-up of AP active site from two angles. Dashes represent putative hydrogen bonds. Residues colored as in part (A). (C) Schematic of AP active site interactions represented with the phosphoryl transfer transition state. Residues colored as in part (A). (D) Reaction scheme for phosphomonoester hydrolysis by AP, where ROP represents a phosphate monoester dianion substrate, and E-P represents the covalent seryl-phosphate intermediate (Coleman, 1992).
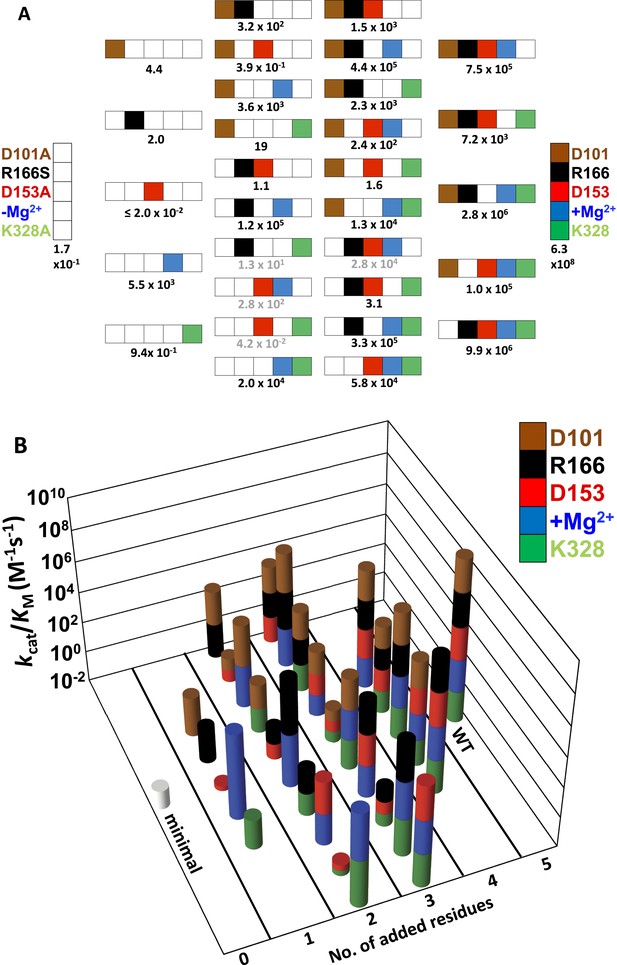
Catalytic efficiencies of AP variants for all combinations of five active site residues.
(A) The 32 possible combinations of the five residues diagrammed with color-coding of residues as in Figure 1 to represent whether a particular WT residue is present: D101, brown; R166, black; D153, red; K328, green; and E322 and the Mg2+ ion, blue; the absence of a WT residue at a particular position is indicated by a white square. The catalytic efficiency, kcat/KM (M−1s−1), of each combination is noted below each construct (Table 1). Rate constants calculated from the energetic behavior of the functional units are shown in grey (Table 1). (B) Three-dimensional representation of the activities of the 32 AP variants, with the height of each bar corresponding to kcat/KM (M−1s−1 on a log scale) and the same color scheme as in (A).
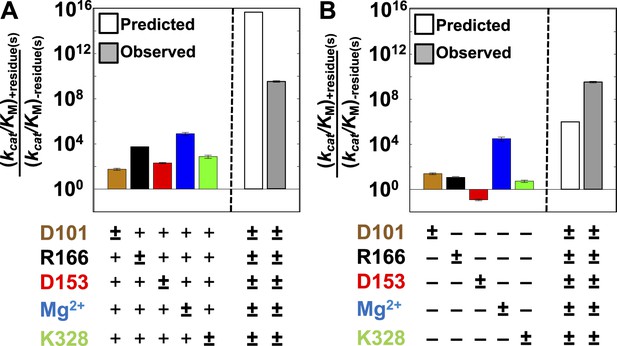
Single-mutation effects and additivity predictions.
Rate effects from removing individual residues from WT AP (A) or restoring individual WT residues to AP minimal (B). The symbol (±) indicates which residue is varied. Residues are color-coded as in Figure 1: D101, brown; R166, black; D153, red; K328, green; and E322 and Mg2+ ion, blue. The following mutations were made: D101A, R166S, D153A, K328A, and E322Y; several alternative mutations gave similar effects (Appendix 1 Table 1). To the right of the dashed line is the activity of WT AP relative to AP minimal observed (A, B, grey bars) and predicted from the effects of removal of each WT residue from the WT background and assuming independent (energetically additive) effects (A, open bar) or from the effects of addition of each WT residue in the minimal background, assuming independence (B, open bar).
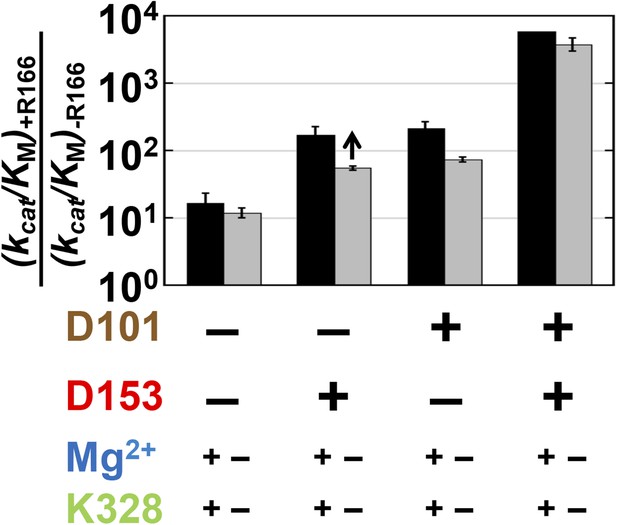
Catalytic effects of R166 in different mutant backgrounds.
The effects of restoring R166 in different aspartate backgrounds with the Mg2+ ion and K328 present (black) or absent (grey). The arrow above the bar in the D153 background indicates that the ratio is a lower limit. Residues are color-coded as in Figure 1, rate constants are from Table 1, and mutations made are listed in Table 1.
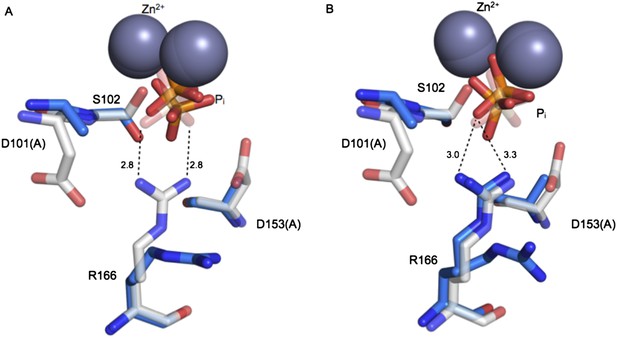
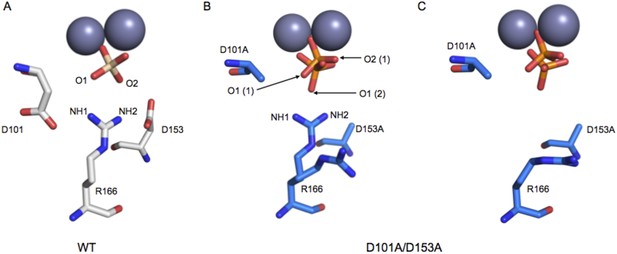
Structure of the active site of D101A/D153A AP.
The active sites of the superimposed crystal structures of Pi-bound D101A/D153A AP (protein: blue, Pi: orange) and Pi-bound WT AP (protein: white, Pi: transparent). The two monomers of the AP dimer exhibit different active site configurations and are therefore both shown (A, B). (In contrast, the WT AP monomers are remarkably similar, as can be seen by comparing panels A and B.) In both monomers of D101A/D153A AP, Pi populates two positions that are distinct from its position in WT AP. (A) In one active site of D101A/D153A, R166 is rotated away from the active site. The hydrogen bond distances between R166 and Pi in WT AP are 2.8 Å (shown in A). (B) In the other active site of D101A/D153A, R166 partially occupies two positions, one of which faces the active site and hydrogen bonds to one of the partially occupied Pi ions. The other rotameric position adopted by R166 is flipped away from the active site, and presumably coincides with the other partially occupied Pi ion, as it would sterically clash with the active-site facing R166 rotamer. Hydrogen bond distances and angles for WT and D101A/D153A AP are listed in Appendix 2 Table 1.
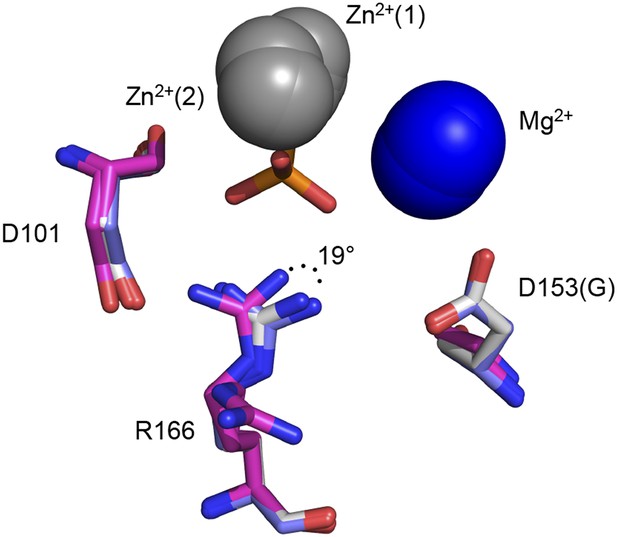
Ablation of D153 disrupts R166 positioning.
Superimposed crystal structures of Pi-bound WT AP (purple, PDB 3TG0), apo WT AP (grey, PDB 1ED9), and apo D153G AP (magenta). In two independent structures of D153G AP, R166 is rotated from its position in Pi-bound WT AP by > 19° (PDB 1AJC) or rotated away from the active site (PDB 1AJD).
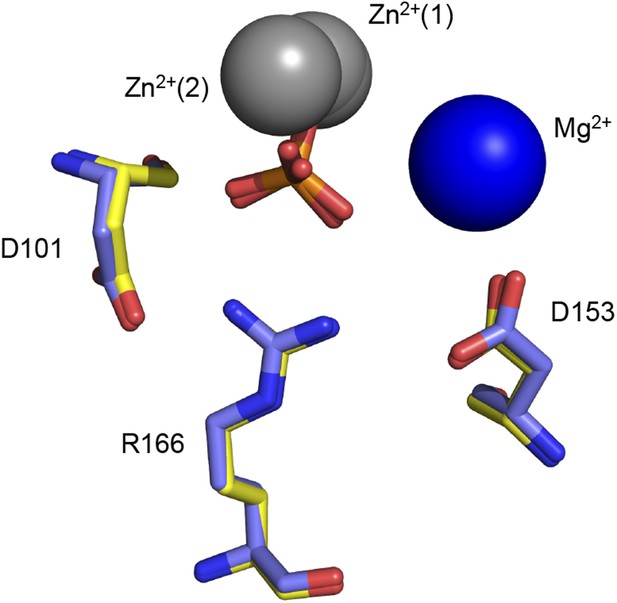
Removal of the active site Mg2+ ion does not disrupt R166 positioning.
Superimposed crystal structures of Pi-bound E322Y AP (yellow, PDB 3DYC), which lacks the active site Mg2+ ion, and Pi-bound WT AP (purple, PDB 3TG0). R166 overlays closely in these structures.
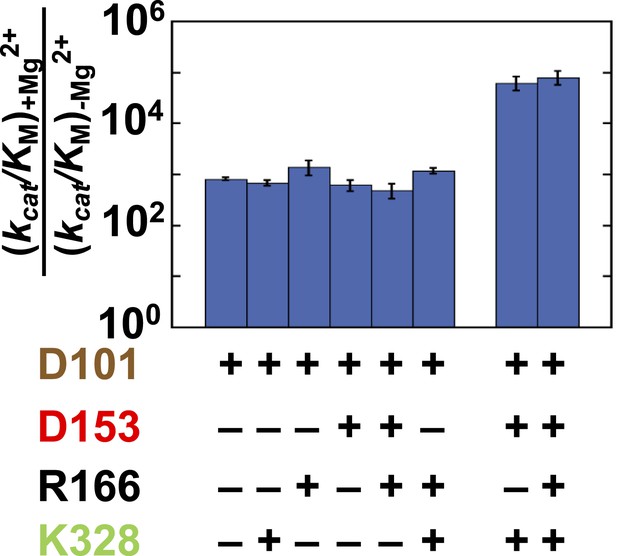
Catalytic effects of the Mg2+ ion in different mutant backgrounds.
The effect of Mg2+ ion addition in different mutational backgrounds. Residues are color-coded as in Figure 1, rate constants are from Table 1, and mutations made are listed in Table 1.
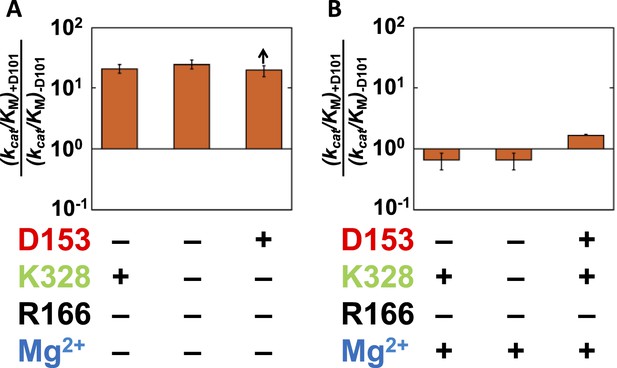
Catalytic effects of D101 in different mutant backgrounds.
The effects of restoring D101 in backgrounds without bound Mg2+ (A) and with bound Mg2+ (B). The arrow indicates that the ratio is a lower limit. Residues are color-coded as in Figure 1, rate constants are from Table 1, and mutations made are listed in Table 1. R166 is absent because it is also coupled with D101 (Figure 4).
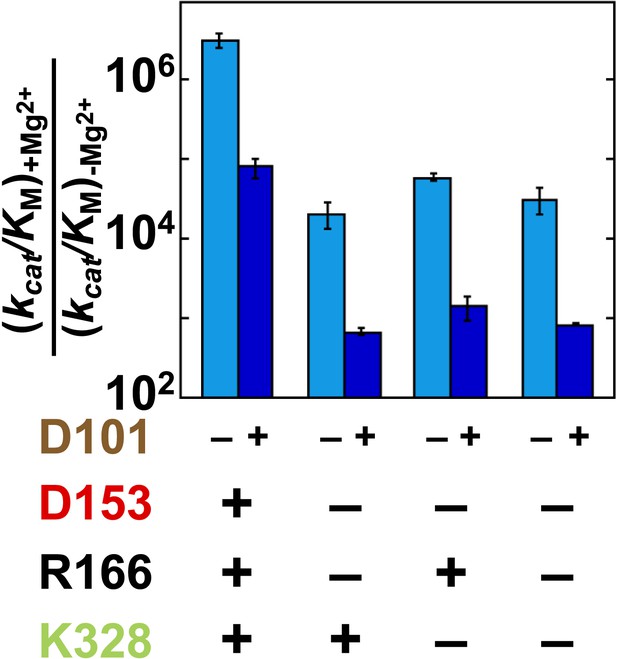
Catalytic effects of Mg2+ ion removal in mutant backgrounds without and with D101 present.
The effects of restoring Mg2+ in the absence of D101 (light blue) and the presence of D101 (dark blue). Residues are color coded as in Figure 1, rate constants are from Table 1, and mutations made are listed in Table 1.
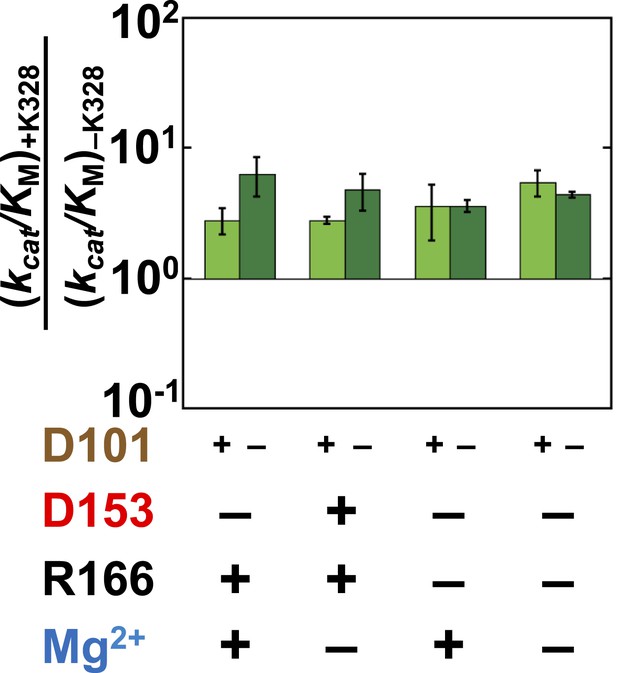
Effect of K328 addition with (light green) or without D101 (dark green) present.
The effect of adding K328 is the same, within approximately twofold, whether D101 is present, indicating that these residues are essentially independent of one another.
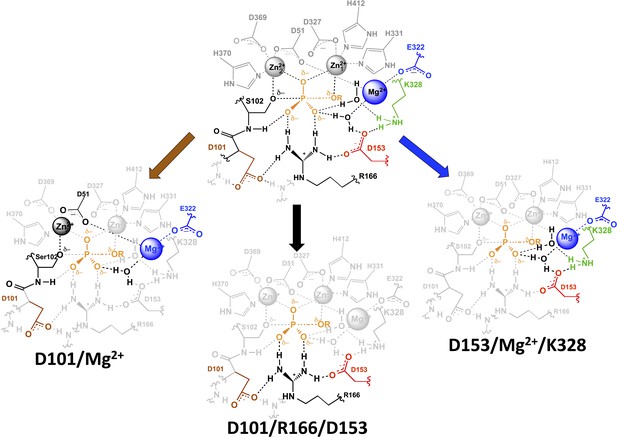
AP active site functional units.
The residues of the functional unit are color-coded as in Figure 1. For the D101/Mg2+ functional unit (left), the black residues and Zn2+ ion represent a potential route for the energetic connections between these residues.
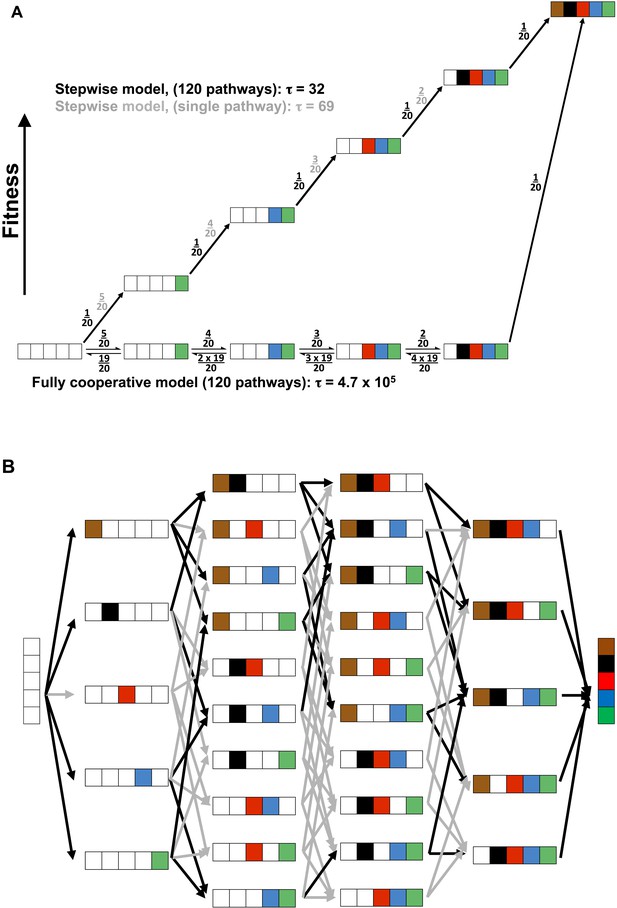
Cooperative and independent models of active site evolution.
(A) Schematic comparing fully cooperative (bottom) and stepwise (top) models for a single pathway. In the fully cooperative model, simultaneous acquisition of all five WT residues is required to confer a selective advantage, leading to a mean waiting time of 4.7 × 105 (arbitrary units) considering all 120 pathways of adding in the residues (black net rates; for simplicity, only one intermediate of the multiple possible mutant combinations is shown in each step). In contrast, the stepwise model, in which acquisition of any WT residue confers a fitness advantage and is thus irreversible (top, black numbers), has a minimum mean waiting time of 32. If only one of the 120 pathways leads to a stepwise increase in fitness (top, grey numbers) then the mean waiting time would be 69. The model and simplifying assumptions made to highlight the differences arising from the presence or absence of cooperativity are described in Appendix 3. (B) Model of active site evolution showing the 120 possible paths in the AP landscape for introduction of the five residues investigated herein, in an otherwise WT background. A stepwise model in which acquisition of any WT residue is considered irreversible and all paths are possible would result in a mean waiting time of 32 (all arrows, grey and black, same as part A, top). As a subset of mutagenic steps toward WT AP (36 of the 80 potential evolutionary steps) confers a selective advantage (here defined as a rate increase of >threefold) and paths containing steps that do not confer such an advantage have much lower probabilities, we consider the 34 of 120 pathways that provide a monotonic fitness increase as all five WT residues are added. This gives a mean waiting time between the mean waiting times for the stepwise models for a single pathway and 120 pathways, 32 and 69, respectively.
Labeling scheme for describing hydrogen bond distances and angles between R166 and Pi.
https://doi.org/10.7554/eLife.06181.022Tables
| AP mutant | kcat/KM (M−1s−1) | fold decrease† | KM (µM) | kcat (s−1) |
|---|---|---|---|---|
| WT | 3.3 × 107‡ | – | 3.6 × 10−1 | 12 |
| (6.3 × 108)§ | (1) | |||
| D101A | 9.9 (2.0) × 106 | 64 | 3.6 (0.9) | 36 (9) |
| R166S¶,# | 1.0 × 105 | 6.3 × 103 | 5.0 | 0.5 |
| D153A | 2.8 (0.4) × 106 | 2.3 × 102 | 2.6 (0.7) | 7.6 (2.8) |
| E332Y**,# | 7.2 (2.2) × 103 | 8.8 × 104 | ∼0.5 | – |
| K328A# | 7.5 (2.4) × 105 | 8.4 × 102 | 5.4 (1.8) | 3.4 (0.9) |
| D101A/R166S | 5.8 (0.2) × 104 | 1.1 × 104 | 8.4 (2.3) | 4.2 (2.0) × 10−1 |
| D101A/D153A# | 3.3 (0.7) × 105 | 1.9 × 103 | 6.2 (2.1) | 2.2 (0.7) |
| D101A/E322Y# | 3.1 (0.1) | 2.0 × 108 | 1.5 (0.3) × 102 | 4.6 (0.8) × 10−4 |
| D101A/K328A†† | (2.8 × 104) | 2.3 × 104 | – | – |
| R166S/D153A | 1.3 (0.1) × 104 | 4.9 × 104 | 5.3 | 7.0 × 10−2 |
| R166S/E322Y** | 1.6 (0.5) | 3.9 × 108 | 27 (11) | – |
| R166S/K328A# | 2.4 (0.4) × 102 | 2.6 × 106 | 6.3 (2.5) × 101 | 1.6 (0.4) × 10−2 |
| D153A/E322Y | 2.3 (0.1) × 103 | 2.7 × 105 | 5.2 (0.4) × 10−1 | 1.4 (0.3) × 10−3 |
| D153A/K328A | 4.4 (1.4) × 105 | 1.4 × 103 | 2.4 | 1.1 |
| E322Y/K328A | 1.5 (0.1) × 103 | 4.2 × 105 | 2.2 | 3.4 × 10−3 |
| D101A/R166S/D153A# | 2.0 (0.7) × 104 | 3.2 × 104 | 4.8 (1.4) | 8.3 (1.8) × 10−2 |
| D101A/R166S/E322Y†† | (4.2 × 10−2) | 1.5 × 1010 | – | – |
| D101A/R166S/K328A†† | (2.8 × 102) | 2.3 × 106 | – | – |
| D101A/D153A/E322Y†† | (1.3 × 101) | 4.8 × 107 | – | – |
| D101A/D153A/K328A | 1.2 (0.1) × 105 | 5.3 × 103 | 11 | 1.6 |
| D101A/E322Y/K328A | 1.1 (0.1) | 5.7 × 108 | 78 | 8.9 × 10−5 |
| R166S/D153A/E322Y | 1.9 (0.1) × 101 | 3.3 × 107 | 4.4 (0.2) × 102 | 8.3 (1.0) × 10−3 |
| R166S/D153A/K328A | 3.6 (0.1) × 103 | 1.8 × 105 | 7.2 (0.1) | 2.6 (0.1) × 10−2 |
| R166S/E322Y/K328A | 3.9 (0.8) × 10−1 | 1.6 × 109 | 1.4 (0.3) × 102 | 5.7 (1.1) × 10−5 |
| D153A/E322Y/K328A | 3.2 (0.3) × 102 | 2.0 × 106 | 2.1 (0.3) | 7.3 (0.3) × 10−4 |
| D101A/R166S/D153A/E322Y | 9.4 (1.6) × 10−1 | 6.7 × 108 | 3.5 × 102 | 3.2 (0.7) × 10−4 |
| D101A/R166S/D153A/K328A | 5.5 (1.7) × 103 | 1.2 × 105 | 12 (5.0) | 6.1 (0.3) × 10−2 |
| D101A/R166S/E322Y/K328A | ≤2.0 × 10−2 | ≥3 × 1010 | – | – |
| D101A/D153A/E322Y/K328A | 2.0 (0.1) | 3.2 × 108 | 1.4 (0.1) × 102 | 2.4 (0.3) × 10−4 |
| R166S/D153A/E322Y/K328A# | 4.4 (0.1) | 1.4 × 108 | 2.2 (0.1) × 102 | 9.6 (0.5) × 10−4 |
| D101A/R166S/D153A/E322Y/K328A# | 1.7 (0.3) × 10−1 | 3.7 × 109 | 1.6 (0.6) × 102 | 2.9 (0.7) × 10−5 |
-
*
Activities were measured in 0.1 M MOPS, pH 8.0, 0.5 M NaCl, 100 µM ZnCl2, and 500 µM MgCl2 at 25°C. Errors in activities are standard deviations from duplicate with the same or independent enzyme preparations (see ‘Materials and methods’).
-
†
‘Fold down’ values are kcat/KM for WT AP estimated for reaction with the chemical step rate limiting (footnote d and description in Appendix 1) divided by (kcat/KM) for each mutant. By definition the value for WT AP is one.
-
‡
(kcat/KM)obsd (O'Brien and Herschlag, 2002).
-
§
For WT AP, the chemical step is not rate limiting (O'Brien and Herschlag, 2002). As has been carried out previously, we used comparisons with a substrate for which the chemical step is rate limiting to estimate the value of kcat/KM for WT AP that would be expected with fast association and the chemical step rate limiting (see description in Appendix 1). Relative values are compared to this number.
-
#
The mutant was expressed in two independent enzyme preparations and standard deviations are from activity measurements for the independent preparations.
-
¶
From reference (O'Brien et al., 2008).
-
**
From reference (Zalatan et al., 2008).
-
††
Values in brackets were not measured but are calculated from the energetic behavior of the functional units according to the mathematical model described in Appendix 1 and shown in Appendix 1 Table 2.
X-ray crystallographic data collection and refinement statistics
| Data collection | |
| Space group | P6322 |
| Unit cell axes | |
| a, b, c (Å) | 161.4, 161.4, 140.1 |
| α, β, γ (ο) | 90.0, 90.0, 120.0 |
| Resolution range (Å) | 52.89–2.24 (2.31–2.24)*,† |
| Rmerge (%) | 43.0 (305.9) |
| Rpim (%)‡ | 9.6 (71.9) |
| <I>/<σI> | 7.8 (1.4) |
| Completeness (%) | 100.0 (100.0) |
| Multiplicity | 21.6 (19.7) |
| CC1/2 | 0.996 (0.582) |
| Refinement | |
| Resolution range (Å) | 52.89–2.24† |
| No. unique reflections | 51,819 (5086) |
| Rwork/Rfree (%) | 21.7/25.9 |
| Number of atoms | 6794 |
| Average B-factors (Å2) | |
| protein | 37.8 |
| water | 30.2 |
| ligands | 41.2 |
| R.m.s. deviation from ideality | |
| Bond length (Å) | 0.006 |
| Bond angles (ο) | 1.10 |
| Ramachandran statistics | |
| Favored regions (%) | 98.4 |
| Allowed regions (%) | 1.6 |
| Outliers (%) | 0 |
| PDB code | 4YR1 |
-
*
Values in parenthesis are for the highest resolution shell.
-
†
The high resolution cut-off applied during scaling and refinement was decided based on CC1/2 and completeness (Diederichs and Karplus, 2013; Evans and Murshudov, 2013).
-
‡
Rpim is reported in addition to Rmerge due to the high multiplicity of the data set.
Kinetic constants for Me-P hydrolysis
| kcat/KM (M−1s−1) | |||
|---|---|---|---|
| AP mutant | pNPP | Me-P | |
| WT*,† | 3.3 × 107 | 1.2 × 106 | 28 |
| 3.5 × 105 | 80 | ||
| R166S† | 1.0 × 105 | 110 | 910 |
| 56 | 1800 | ||
| E322Y‡ | 7.2 × 103 | 1.6 | 4500 |
| D101A | 9.9 × 106 | 2.7 × 103 | 3600 |
| D153A | 2.8 × 106 | 1.1 × 103 | 2500 |
| D101A/D153A | 3.3 × 105 | 61 | 5400 |
-
*
(kcat/KM)obsd; the chemical step is not rate limiting.
-
†
Values of kcat/KM of 1.2 × 106 M−1s−1 and 110 M−1s−1 for WT and R166S, respectively for Me-P was obtained previously from reference (O'Brien and Herschlag, 2002; O'Brien et al., 2008). This difference in value would not affect the conclusions herein.
-
‡
From reference (Zalatan et al., 2008).
-
The efficiency of Me-P hydrolysis was measured for mutants with pNPP activities close to the rate of diffusion. The ratios measured for the enzyme with pNPP and Me-P were close to what had been measured previously for R166S and E322Y, two mutants for which chemistry is rate limiting for both substrates. The ratios suggest that chemistry is rate limiting for the pNPP hydrolysis in D101A and D153A.
Alternative mutations
| AP mutant | kcat/KM (M−1s−1) | krel |
|---|---|---|
| D101A | 9.9 (2.0) × 106 | (1) |
| D101A* | 1.0 × 107 | 1.0 |
| D101G | 7.6 (1.3) × 106 | 1.5 |
| D101S | 9.1 (1.3) × 106 | 1.1 |
| D101S† | 3.1 × 106 | 3.2 |
| D101S‡ | 1.7 × 106 | 5.8 |
| D153A | 2.8 (0.4) × 106 | (1) |
| D153A§ | 1.5 × 106 | 1.9 |
| D153G‡ | 1.6 × 106 | 1.8 |
| R166S# | 1.0 × 105 | (1) |
| R166S¶ | 5.8 × 104 | 1.7 |
| R166A# | 2.0 × 104 | 5.0 |
| E332Y** | 7.2 (2.2) × 103 | (1) |
| E322A** | 8.9 × 103 | 0.8 |
| K328A** | 7.5 (2.4) × 105 | (1) |
| K328A†† | 3.6 × 106 | 0.2 |
| R101A/R166S | 5.8 (0.2) × 104 | (1) |
| R101G/R166S | 2.7 (0.3) × 104 | 2.1 |
| R166S/D153A/E322Y/K328A | 4.4 (0.1) | (1) |
| R166S/D153A/E322A/K328A | 2.7 | 1.6 |
| D101A/R166S/D153A/E322Y/K328A | 1.7 (0.3) × 10−1 | (1) |
| D101G/R166S/D153A/E322Y/K328A | 1.3 (0.2) × 10−1 | 1.3 |
| D101N/R166S/D153A/E322Y/K328A | 1.9 (0.3) × 10−1 | 0.9 |
-
*
Reference (Herschlag, 1988).
-
†
Reference (Freedman et al., 2009).
-
‡
Reference (Herschlag, 1991).
-
§
Reference (Wolfenden, 1976).
-
#
Reference (Eisenmesser et al., 2002).
-
¶
Reference (Lanzetta et al., 1979).
-
**
Reference (Halabi et al., 2009).
-
††
Reference (Battye et al., 2011).
Comparison of predicted and observed catalytic efficiencies of the 32 possible AP mutants
| AP mutant | (kcat/KM)Predicted (M−1s−1) | Ratio measured/predicted |
|---|---|---|
| WT | 4.2 × 108 | 1.5 |
| D101A | 9.7 × 106 | 1.0 |
| R166S | 8.5 × 104 | 1.2 |
| D153A | 1.9 × 106 | 1.5 |
| E332Y | 6.6 × 103 | 1.1 |
| K328A | 1.2 × 106 | 0.6 |
| D101A/R166S | 9.6 × 104 | 0.6 |
| D101A/D153A | 3.9 × 105 | 0.8 |
| D101A/E322Y | 4.2 | 0.7 |
| D101A/K328A* | 2.8 × 104 | N/A |
| R166S/D153A | 1.9 × 104 | 0.7 |
| R166S/E322Y | 1.3 | 1.2 |
| R166S/K328A | 2.5 × 102 | 1.0 |
| D153A/E322Y | 2.2 × 103 | 1.0 |
| D153A/K328A | 4.1 × 105 | 1.1 |
| E322Y/K328A | 1.4 × 103 | 1.1 |
| D101A/R166S/D153A | 2.1 × 104 | 0.7 |
| D101A/R166S/E322Y* | 4.2 × 10−2 | N/A |
| D101A/R166S/K328A* | 2.8 × 102 | N/A |
| D101A/D153A/E322Y* | 13 | N/A |
| D101A/D153A/K328A | 8.5 × 104 | 1.4 |
| D101A/E322Y/K328A | 0.92 | 1.2 |
| R166S/D153A/E322Y | 22 | 0.9 |
| R166S/D153A/K328A | 4.0 × 103 | 0.9 |
| R166S/E322Y/K328A | 0.29 | 1.3 |
| D153A/E322Y/K328A | 4.8 × 102 | 0.7 |
| D101A/R166S/D153A/E322Y | 0.68 | 1.4 |
| D101A/R166S/D153A/K328A | 4.5 × 103 | 1.2 |
| D101A/R166S/E322Y/K328A | 9.0 × 10−3 | 2.2† |
| D101/D153A/E322Y/K328A | 2.8 | 0.7 |
| R166S/D153A/E322Y/K328A | 4.8 | 0.9 |
| D101A/R166S/D153A/E322Y/K328A | 0.15 | 1.1 |
-
*
Obtained from the predicted values in this table and the observed values in Table 1.
-
†
Only an upper limit for this mutant could be measured.
| AP variant | Donor–Acceptor‡ | Distance* (Å) | Angle† (°) |
|---|---|---|---|
| WT | NH1-O1 | 2.8 | 170 |
| NH2-O2 | 2.8 | 177 | |
| D101A/D153A | NH1-O1 (1) | 3.0 | 158 |
| NH2-O1 (1) | 3.4 | 139 | |
| NH1-O2 (1) | 4.9 | 131 | |
| NH2-O2 (1) | 4.3 | 142 |
-
*
Distances between donor and acceptor atoms.
-
†
The angle between the donor, predicted hydrogen, and acceptor atoms.
-
‡
NH1 and NH2 refer to the nitrogen atoms of the guanidinium group of R166. O1 and O2 refer to the relevant oxygen atom of the bound Pi; in the case of D101A/D153A, the number in parenthesis refers to the partially occupied Pi labeled in the Appendix 2 Figure 1 below. Only distances and angles to one of these phosphate ions in the monomer shown in Appendix 2 Figure 1 panel (B) are listed in the table; the other Pi sterically clashes with the active-site facing rotamer of R166 and thus presumably is populated only when the R166 rotamer is flipped away from the active site. Distances and angles are not reported for the other subunit (shown in Appendix 2 Figure 1C below) because in this active site, R166 is flipped out of the active site so that it is not within hydrogen bonding distance of the bound Pi. For reference, the WT active site is shown in Appendix 2 Figure 1 panel (A) below.





