Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other
Figures
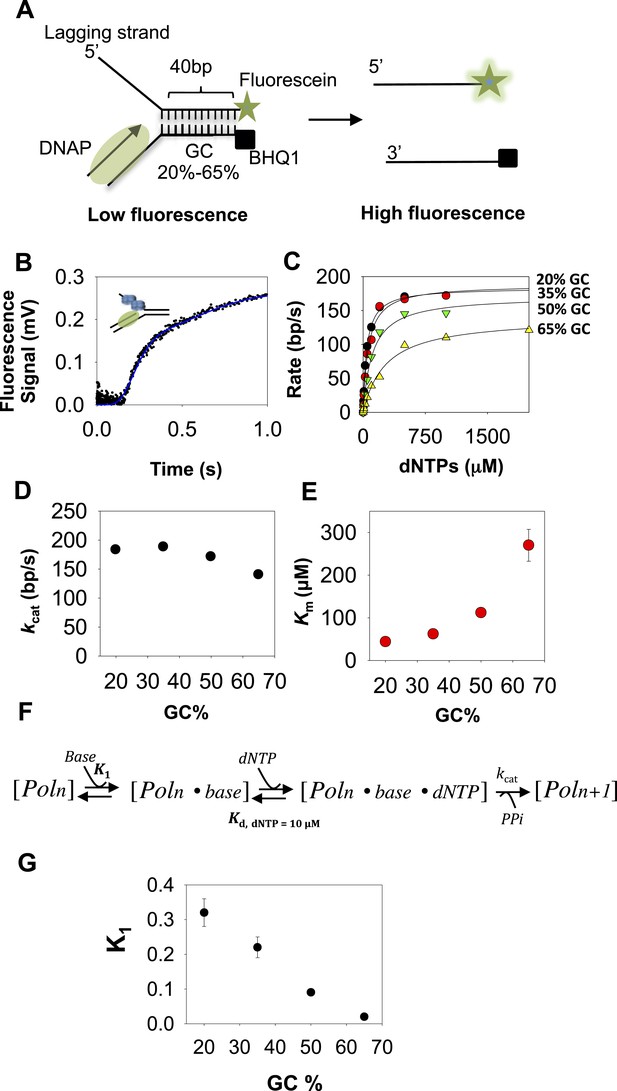
Kinetic mechanism of DNA unwinding by T7 DNAP.
(A) Preformed replication fork DNA substrate for the measurement of the unwinding kinetics of T7 DNA polymerase (DNAP). (B) Representative kinetics of DNA unwinding by T7 DNAP in the presence of E. coli single-stranded DNA-binding protein (SSB) (dots) fit to the n-step model (solid line). (C) The base pair unwinding rates are plotted against dNTPs concentration and fit to Equation 4 (solid line) to obtain the unwinding kcat and dNTPs Km values. (D, E) The unwinding kcat and Km of T7 DNAP plotted against GC percentages. (F) The three-step ordered model of base-capture, dNTP binding, and nucleotide incorporation that describes the unwinding kinetics of T7 DNAP. (G) Equilibrium constants of the base-capture step obtained from fitting the kinetics data to the three-step model in F are plotted against increasing GC percentages. Details of the kinetic fittings are provided in Appendix—Section 2.
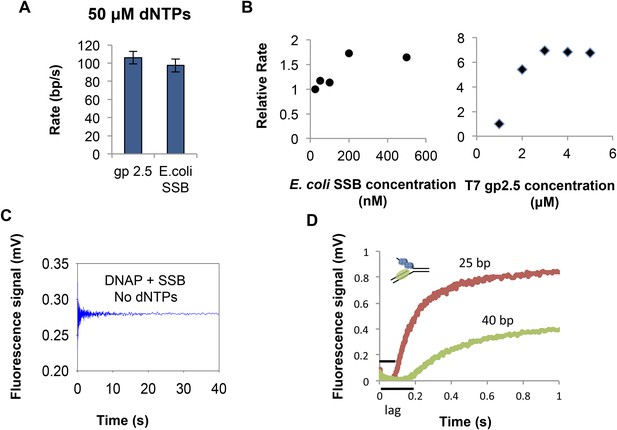
Unwinding by T7 DNAP with gp2.5 and SSB.
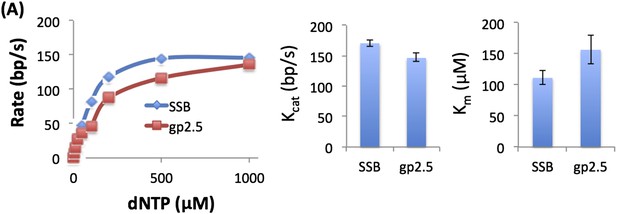
(A) Unwinding rate of T7 DNAP with T7 gp2.5 (3 μM) and E. coli SSB (200 nM) of the 20% GC DNA substrate at 50 μM dNTPs concentration. Both T7 gp2.5 and E. coli SSB can stimulate the unwinding rate of the T7 DNAP. (B) Relative rate of DNA unwinding by T7 DNAP with increasing concentration of E. coli SSB to determine the optimum concentration of SSB to use in the unwinding experiments. ‘SSB concentration’ refers to monomer concentrations. Relative rate of DNA unwinding of T7 DNAP with increasing concentration of T7 gp2.5 shows that stimulation by T7 gp2.5 saturates at ∼3 μM concentrations. (C) No increase in fluorescence signal from the DNA (20% GC) is observed in the absence of DNA synthesis by the T7 DNAP showing that E. coli SSB by itself does not unwind the DNA. (D) Kinetic trace for strand displacement DNA synthesis using the fluorescence-based assay shows the increase in lag time when the length of the dsDNA in the replication fork substrate is increased. The experiment was done in a stopped-flow setup with 35% GC DNA substrate and 100 μM dNTPs concentration.
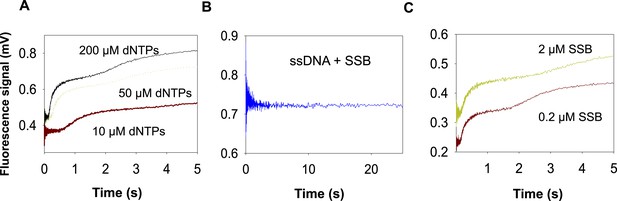
Unwinding trace of T7 DNAP with SSB.
(A) The kinetic trace for strand displacement DNA synthesis using the fluorescence-based assay shows an initial time lag followed by fluorescence increase in two phases. The traces shown are using the 20% GC DNA substrate. (B) Effect of SSB binding to ssDNA with fluorescein in the 3′ end of the ssDNA. A description of the kinetic trace is provided in Appendix—Section 1. (C) Effect of increasing SSB concentration on the second phase observed in the fluorescence traces obtained with DNAP and SSB. The traces shown are using the 35% GC DNA substrate.
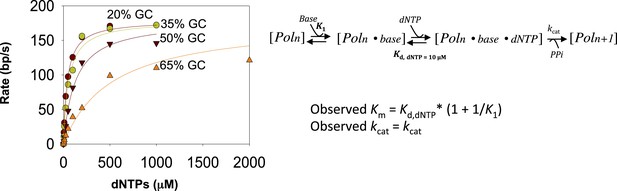
Fitting kinetics of unwinding by T7 DNAP with SSB.
The rate dependencies of the T7 DNAP as a function of dNTPs concentration for dsDNA of increasing GC content were fit to the minimal mechanism shown above to estimate the K1 values shown in Figure 1G. Kd, dNTP was fixed at 10 μM, which corresponds to the Kd, dNTP for the DNAP on primer-template DNA. kcat was allowed to float as a global parameter. Details about the fitting equations are provided in Appendix—Section 2.
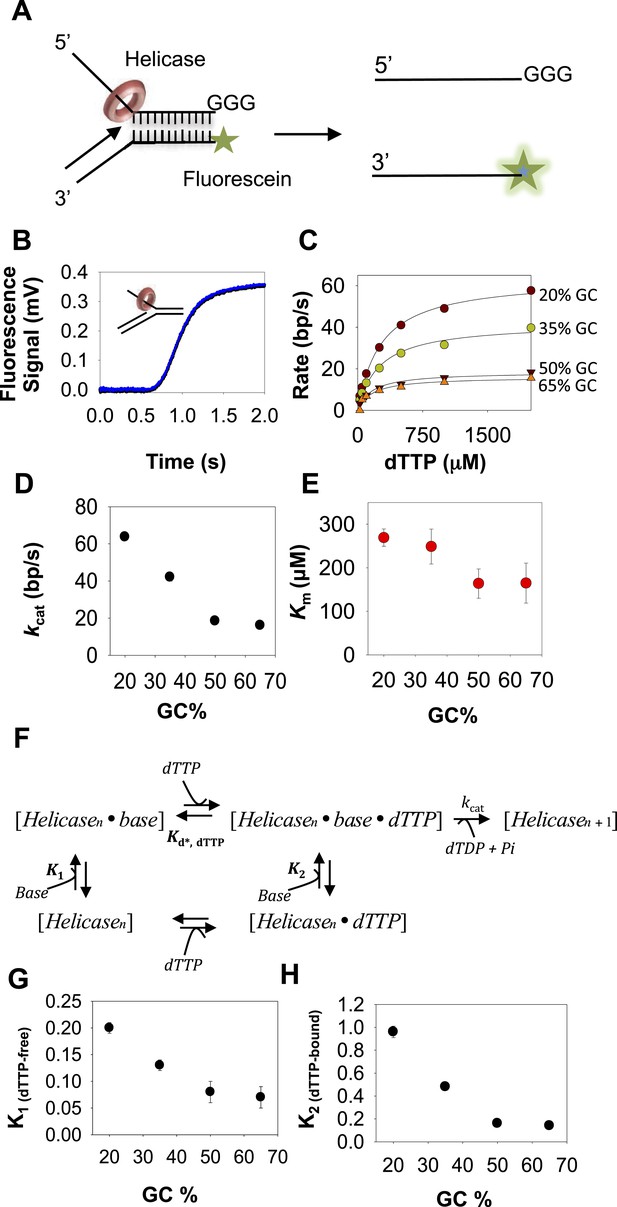
Kinetic mechanism of DNA unwinding by T7 helicase.
(A) Replication fork DNA for the measurement of the unwinding kinetics of T7 helicase. (B) Representative kinetic trace of DNA unwinding by T7 helicase (dots) fit to the n-step model (solid line). (C) The unwinding rates against dTTP concentrations fit to Equation 4 (solid line) to obtain the unwinding kcat and dTTP Km values. (D, E) The kcat and dTTP Km plotted as a function of GC percentages. (F) Schematic of the helicase model with random order of base-capture and dTTP binding. (G, H) Plots of equilibrium constants for the base-capture steps in the dTTP free (K1) and dTTP bound (K2) state as a function of GC percentages. Kd, dTTP was fixed at 90 μM, which corresponds to the Km, dTTP for the helicase when translocating on ssDNA (Figure 2—figure supplement 2 and Appendix—Section 4). kcat was fixed at 130 nt/s corresponding to the ssDNA translocation rate of the helicase (Kim et al., 2002). Details of the fittings are provided in Appendix—Section 3.
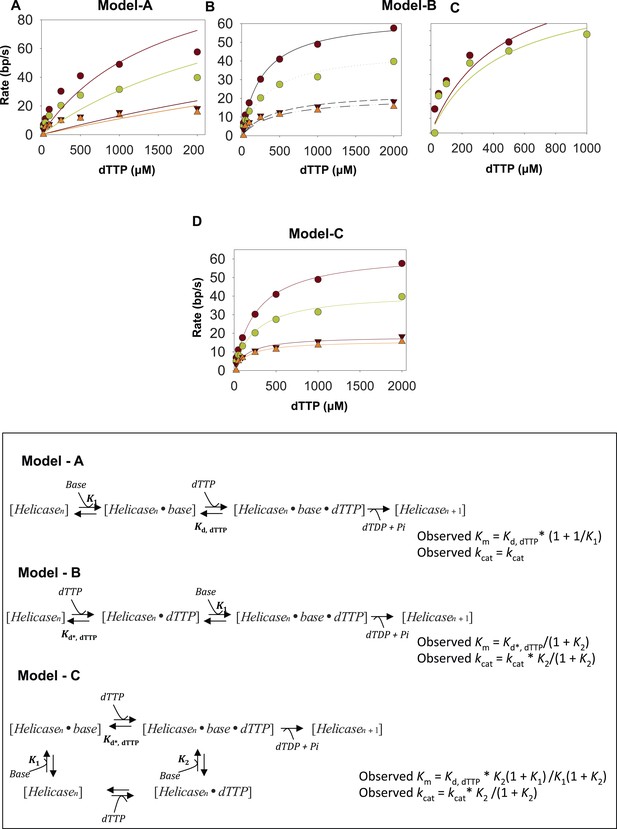
Fitting kinetics of unwinding by T7 helicase.
The dTTP dependency data for unwinding by the helicase on the 20–65% GC DNA were fit to the model A (A) model B (B and C) and model C (D). The data do not fit well to model A. Although it fits reasonably well to model B for the 20% and 35% GC DNA, the 50% and 65% GC DNA data do not fit well to the model. (C) The best fits were obtained with model C. (D) The fitting was performed using SigmaPlot. The derivation of equations used in the fitting is shown in the Appendix—Section 3. The Km, dTTP was fixed at 90 μM in the fitting based on the Km, dTTP for the helicase when translocating on ssDNA substrate (Appendix—Section 4). The kcat was fixed at 130 nt/s corresponding to the ssDNA translocation rate of the helicase.
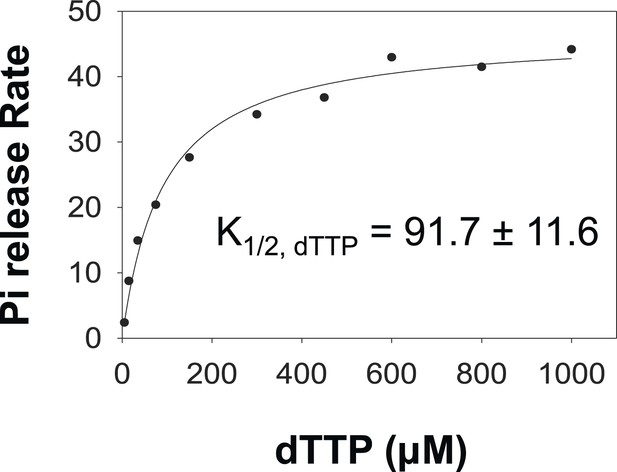
Pi release kinetics.
Burst Rate of Pi release plotted as a function of dTTP to obtain the Km, dTTP in the presence of dT90 ssDNA. The error represents fitting error.
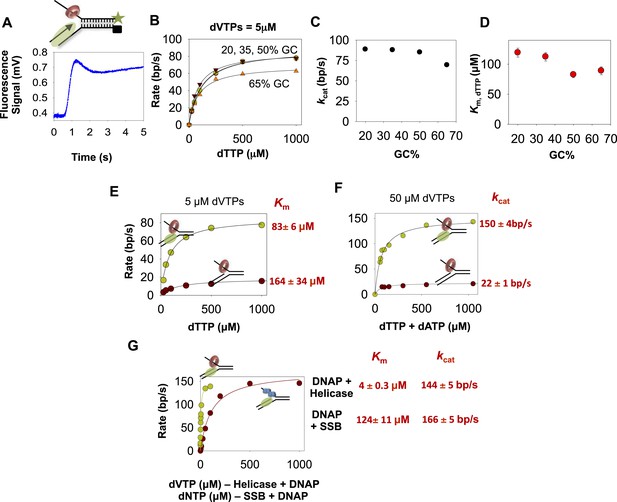
The kinetics of DNA unwinding by the combined helicase and DNAP enzymes.
(A) The replication fork DNA substrate and representative kinetic trace of DNA unwinding by the combined T7 DNAP and T7 helicase enzymes. (B) The base pair unwinding rates of the combined enzymes at 5 µM dVTPs plotted against dTTP concentrations and fit to Equation 4 (solid lines) to obtain the maximal rate of unwinding (kcat) and Km for dTTP. (C, D) The unwinding kcat and dTTP Km as a function of GC percentages. (E) The unwinding rates of the isolated helicase (red circles) and helicase-DNAP (green circles) were measured using the 50% GC fork at constant 5 μM dVTP and increasing dTTP concentrations. (F) The unwinding rates of the isolated helicase (red circle) and helicase-DNAP (green circle) were measured using the 50% GC fork at 50 μM dVTP concentrations and increasing dTTP concentrations. (G) The unwinding rates of T7 DNAP with E. coli SSB (red circle) or with T7 helicase (green circle) were measured using the 50% GC fork at 500 μM dTTP and increasing dNTPs concentrations.
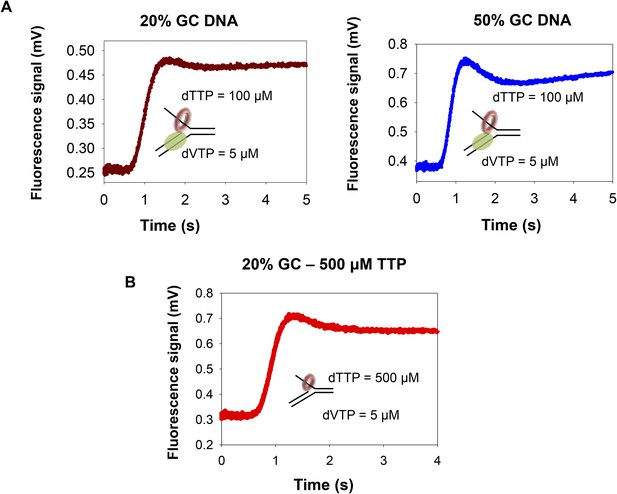
Unwinding-synthesis trace for helicase-DNAP.
(A) Representative trace showing time course of unwinding-synthesis by the helicase–polymerase on the 20% GC and 50% GC DNA substrate at 100 μM dTTP and 5 μM dVTP. The dip in signal is more prominent with the 50% GC DNA substrate compared to the 20% GC DNA substrate. This suggests that the first and second phase could represent the coupled and uncoupled activities of the helicase-DNAP, respectively. (B) Representative trace showing time course of unwinding by the helicase at 500 μM dTTP on the 20% GC DNA. The presence of the dip in the helicase only reaction suggests that interaction of helicase with the fluorescein leads to a dip in the fluorescence signal once the strands are unwound. Together, these data indicate that the dip in the fluorescence signal could be due to (i) helicase interaction with the fluorescein and (ii) coupled and uncoupled activities of the helicase-DNAP.
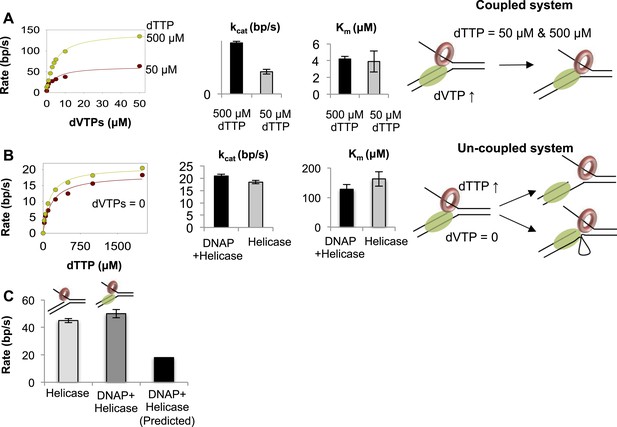
Functionally coupled and uncoupled helicase-DNAP.
(A) The unwinding rates of the combined helicase-DNAP were measured at 50 µM dTTP (red circle) or 500 µM dTTP (green circle) at increasing dVTPs concentration on the 50% GC fork. The bar chart shows the unwinding kcat and dVTPs Km of the combined enzymes at low- (grey bars) and high-dTTP (black bars) concentrations. The cartoon shows that the enzymes remain functionally coupled when helicase is the slow motor. (B) The unwinding rates of the combined enzymes (green circles) at zero dVTPs concentration are compared to the rates of helicase alone (red circles) at increasing dTTP concentrations on the 50% GC fork. The bar charts compare the unwinding kcat and dTTP Km of helicase-DNAP (black) and isolated helicase (gray). Error bars represent fitting errors. The cartoon show that stalling DNAP leads to functional uncoupling between helicase-DNAP with or without physical uncoupling. (C) The DNA unwinding rates of the isolated helicase and helicase-DNAP were measured at 1 mM dTTP and 0.5 µM dVTPs on the 20% GC fork. The bar chart shows the unwinding rate of the isolated helicase (light gray), helicase-DNAP complex (dark gray), and the predicted rate of DNA synthesis by helicase-DNAP assuming coupled synthesis (black).
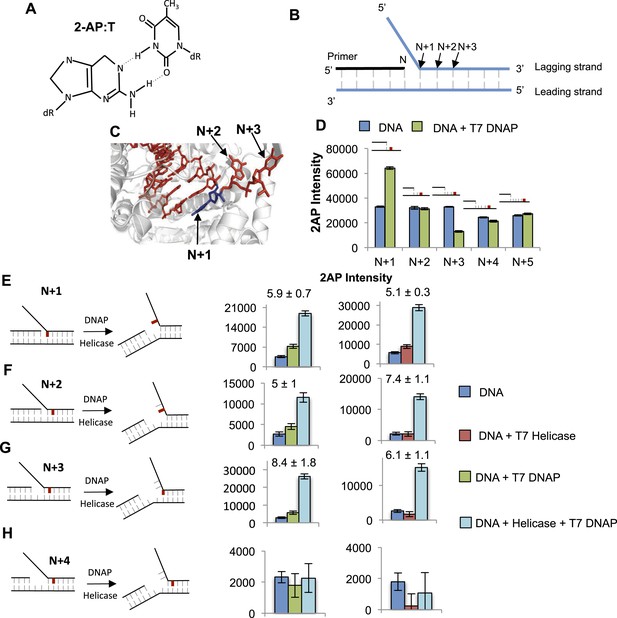
Base pair melting by isolated and combined T7 DNAP and T7 helicase using 2-aminopurine fluorescence changes.
(A) Structure of the 2-aminopurine (2-AP):T base pair. (B) Structure of the replication fork substrate for 2-AP studies. The primer-end is N and subsequent base pairs are N + 1, N + 2, etc. The substrates contained a single 2-AP in the leading or the lagging strand. The primer-end is next to the junction base pair as shown or separated by gaps of one to three template strand nucleotides (not shown). (C) Crystal structure of T7 DNAP bound to primer-template DNA substrate (PDB: 2AJQ). The N + 1 base (blue) is bound in the insertion site, and the N + 2 and N + 3 bases are bound in the template-binding channel. The figure was made using PyMOL (Schrodinger, 2010). (D) Fluorescence intensities of 2-AP modified primer-template substrate without (blue) and with T7 DNAP (green). The 2-AP probe is shown in red at the indicated positions. (E–H) Fluorescence intensities of 2-AP modified replication fork substrates with and without T7 DNAP and T7 helicase. The cartoons show the structure of fork DNA before and after binding of combined T7 DNAP and T7 helicase enzymes. Errors shown are standard deviations from average of 2–5 experiments.
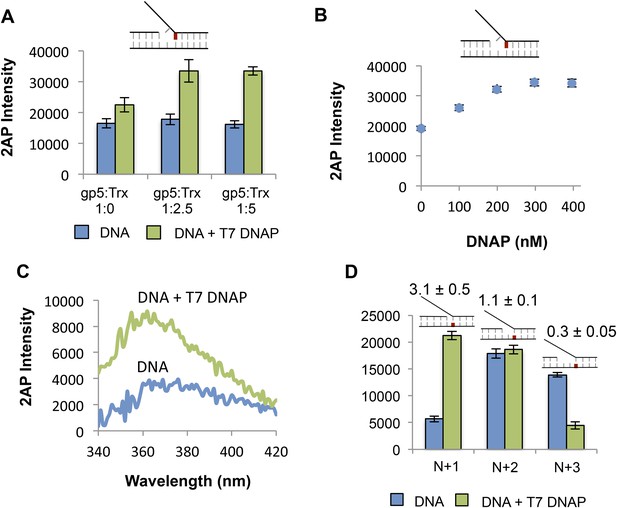
Determining optimal conditions for T7 DNAP and extent of influence of T7 DNAP on template bases in replication fork substrate.
(A) Effect of increasing ratio of the T7 DNAP processivity factor thioredoxin on the 2-AP fluorescence signal. A final ratio of 1:2.5 was used in all the experiments. (B) Effect of DNAP concentration on fluorescence emission of 2-AP in the +1 lag 1 gap substrate. Final enzyme concentration of 200 nM was used in all experiments. (C) Sample fluorescence intensity trace showing the effect of T7 DNAP binding to No gap replication fork DNA. The 2-AP intensity at 370 nm was used in all experiments. (D) Fluorescence intensities of replication fork DNAs with 2-AP at various indicated positions in the leading strand with and without T7 DNAP.
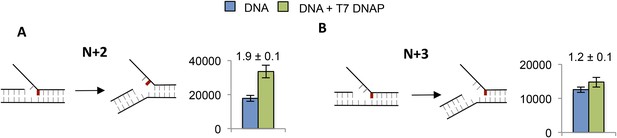
Effect of T7 DNAP on 2-AP at the junction at N + 2 and N + 3.
(A and B) Fluorescence intensities of fork DNA with 2-AP part of the junction base pair at A. N + 2 and B. N + 3 positions in the lagging strand with and without T7 DNAP. The cartoons show the structure of fork DNA before and after binding of T7 DNAP.
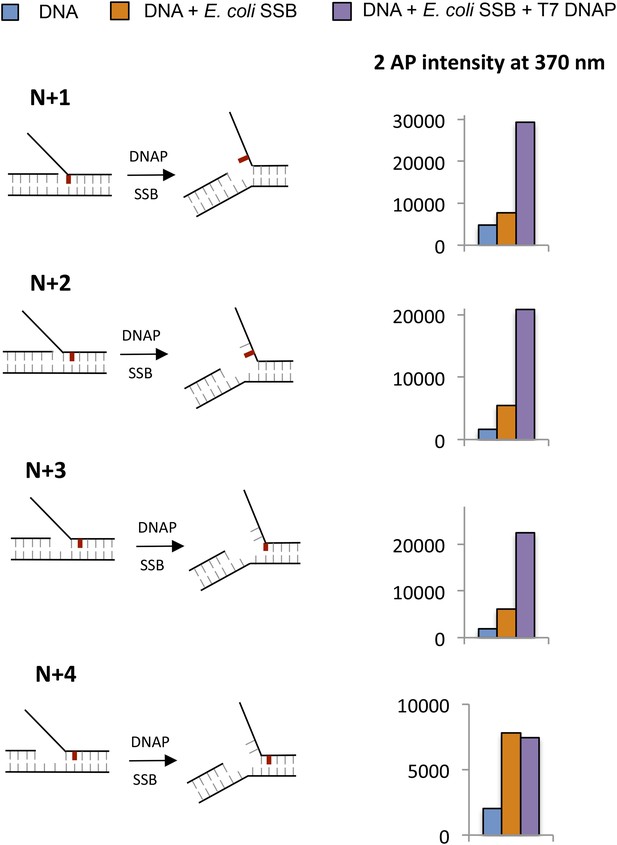
Base pair melting by combined SSB—DNAP using 2-AP fluorescence change.
Fluorescence intensities of 2-AP modified replication fork substrates with and without T7 DNAP and E. coli SSB. The cartoons show the structure of fork DNA before and after binding of combined T7 DNAP and E. coli SSB enzymes. Each experiment had 100 nM DNA, 200 nM DNAP, and 200 nM SSB (tetramer).
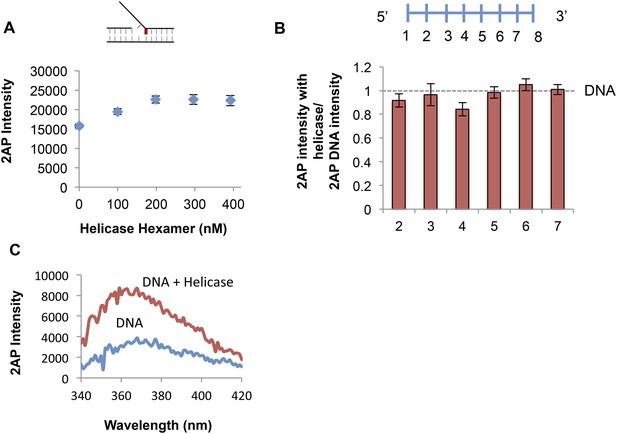
Determining optimal conditions for T7 helicase.
(A) Effect of helicase concentration on fluorescence emission of 2-AP in the +1 lag 1 gap substrate. Final enzyme concentration of 200 nM was used in all experiments. (B) Fold change on helicase binding to short 8 nt ssDNA substrates with probe at positions 2, 3, 4, 5, 6, and 7 from the 5′ end. Error bars represent instrument fluctuations. (C) Sample fluorescence intensity trace showing the effect of helicase binding to No gap replication fork DNA. The 2-AP intensity at 370 nm was used in all experiments.
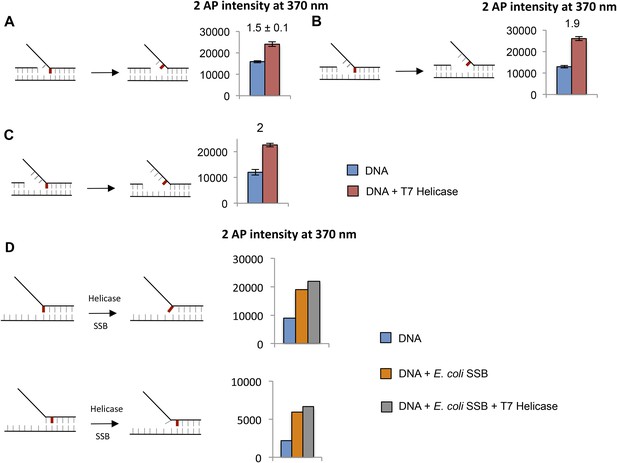
(A–C) Fluorescence intensities of fork DNA with 2-AP in the lagging strand at the fork junction and increasing distance between primer end and fork junction with (red bars) and without T7 helicase (blue bars).
The cartoons show the structure of fork DNA before and after binding of T7 helicase. The numbers above the bar refer to fold change of 2-AP intensity on protein binding to the DNA. (D) Fluorescence intensities of 2-AP modified fork substrates with and without T7 Helicase and E. coli SSB. The cartoons show the structure of fork DNA before and after binding of combined T7 Helicase and E. coli SSB enzymes. Fork substrates with two single-stranded DNA overhangs were used in the experiments with SSB and helicase to provide a binding site for both enzymes on opposite strands.
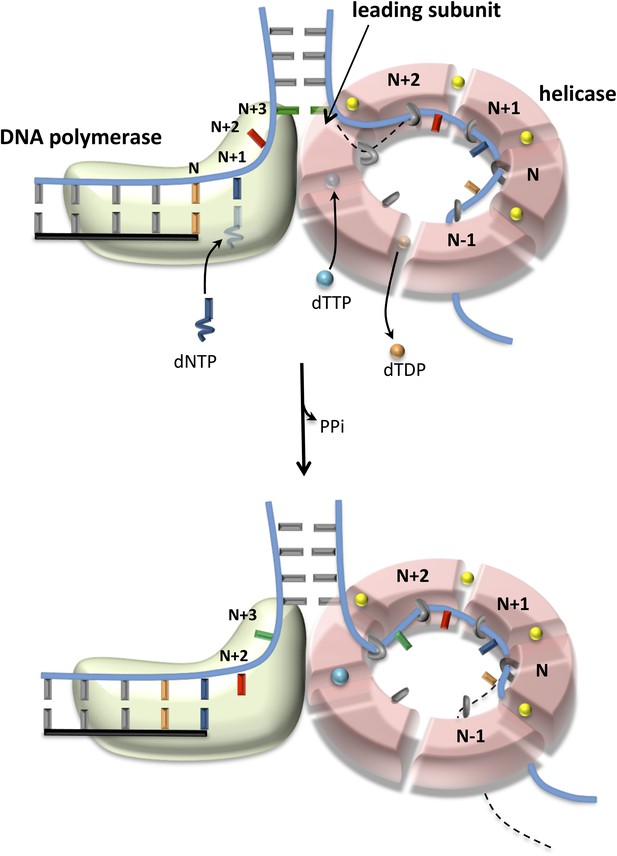
Proposed model of DNA unwinding-synthesis by T7 replisome.
The top cartoon of T7 replisome results after melting of the N + 2 base pair by helicase and DNAP. There are two unwound nucleotides (N + 1 and N + 2) between the primer-end and fork junction at N + 3. The N + 1 base of the leading strand is bound in the insertion site and serves as the templating nucleotide for the incoming dNTP, and the N + 2 is unstacked and bound in the template-binding channel of the DNAP. The complementary N + 1 and N + 2 nucleotides (as well as N and N − 1) on the lagging strand are bound to individual subunits of the helicase hexamer, as shown. Stable binding to N + 2 base by the helicase triggers dTTP hydrolysis and products release at different subunits of the ring. The helicase subunit at the leading edge has weak interactions with the partially unwound N + 3 junction base (dotted line), which gets stabilized after the next round of catalysis. When N is elongated by one nucleotide, the N + 2 moves into the insertion site after PPi release, and the helicase and DNAP cooperatively melt the N + 3 junction base pair, as shown in the bottom cartoon. This model explains the one-nucleotide step size where the combined enzymes translocate by one nucleotide for every dTTP hydrolyzed and nucleotide incorporated (Pandey and Patel, 2014).
Additional files
-
Supplementary file 1
DNA sequences used in unwinding and 2-aminopurine experiments.
- https://doi.org/10.7554/eLife.06562.020





