Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments
Figures
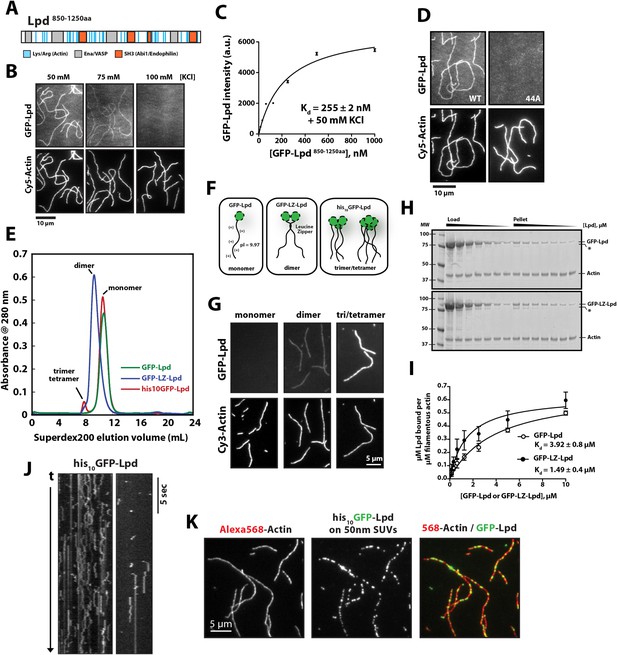
Lamellipodin (Lpd) binds directly to single actin filaments in vitro.
(A) Cartoon representation of the human Lpd850−1250aa highlighting the Enabled/Vasodilator (Ena/VASP) binding sites (grey), Abi1/endophilin SH3 binding sites (red), and basic amino acid residues comprising the actin-binding region (blue). (B) Representative Total Internal Reflection Fluorescence (TIRF)-M images showing 500 nM monomeric GFP-Lpd850−1250aa bound to single actin filaments in the presence of TIRF buffer containing 20 mM HEPES [pH 7.0], 50–100 mM KCl, 1 mg/ml BSA, and 1 mM TCEP. Scale bar, 10 µm. (C) Calculation of Kd for GFP-Lpd850−1250aa actin filament binding using the average fluorescence intensity of GFP-Lpd bound to phalloidin stabilized actin filaments (20% Cy5 labeled). Error bars represent standard error of the mean. (D) Mutations in all 44 lysine/arginine residues to alanine (called Lpd44A) abolish F-actin binding of GFP-Lpd850−1250aa. Actin filament binding was visualized in the presence 500 nM GFP-Lpd850−1250aa, wild-type and 44A mutant, in the presence of 50 mM KCl containing buffer as in (B). Scale bar, 10 µm. (E) Purification of GFP-Lpd850−1250aa (monomer), GFP-LZ-Lpd850−1250aa (dimer), and his10-GFP-Lpd850−1250aa (monomers and trimer/tetramer) by size exclusion chromatography. (F) Cartoon representation of purified Lpd oligomers in (E). (G) Oligomerization of GFP-Lpd850−1250aa enhances actin filament binding. Localization of 250 nM GFP-Lpd850−1250aa (monomer), GFP-LZ-Lpd850−1250aa (dimer), and his10-GFP-Lpd850−1250aa (oligomers) bound to phalloidin stabilized actin filaments (20% Cy5 labeled) in the presence of TIRF buffer containing 100 mM KCl. Scale bar, 5 µm. (H) Representative SDS-PAGE showing co-sedimentation of 1 µM filamentous actin in the presence of increasing concentrations of GFP-Lpd or GFP-LZ-Lpd (0–10 µM monomer concentration). Asterisks (*) on SDS-PAGE gel marks partially translated or proteolyzed GFP-Lpd and GFP-LZ-Lpd that could not be removed during the purification. (I) Calculation of Kd for GFP-Lpd and GFP-LZ-Lpd actin binding domains (BDs) by actin co-sedimentation in the presence of 100 mM KCl buffer (± represents error of fit; error bars are S.D. of the mean from two independent experiments). Note that a small fraction of Lpd is non-specifically absorbed to the walls of the centrifuge tubes in the actin co-sedimentation assay. As a result, the stoichiometry of Lpd bound to actin is likely over-estimated by 5–10% (see ‘Materials and methods’). (J) Kymograph showing diffusion of his10-GFP-Lpd850−1250aa oligomers along the length of a phalloidin stabilized actin filaments. Vertical scale bar, 5 s. (K) Membrane bound his10GFP-Lpd850−1250aa associates with single actin filaments. Localization of 50 nm extruded small unilamellar vesicles (SUVs) DOPC/DOGS-NTA(Ni+2) (99:1 molar ratio) coated with his10GFP-Lpd850−1250aa bound to Alexa568 phalloidin stabilized actin filaments. 25 nM his10GFP-Lpd850−1250aa from (E) was combined with 50 nm SUVs (5 µM total lipid containing 1% or 50 nM DOGS-NTA lipid) in buffer containing 20 mM HEPES [pH7], 100 mM KCl, 100 µg/ml BSA, 1 mM TCEP. Scale bar, 5 µm.
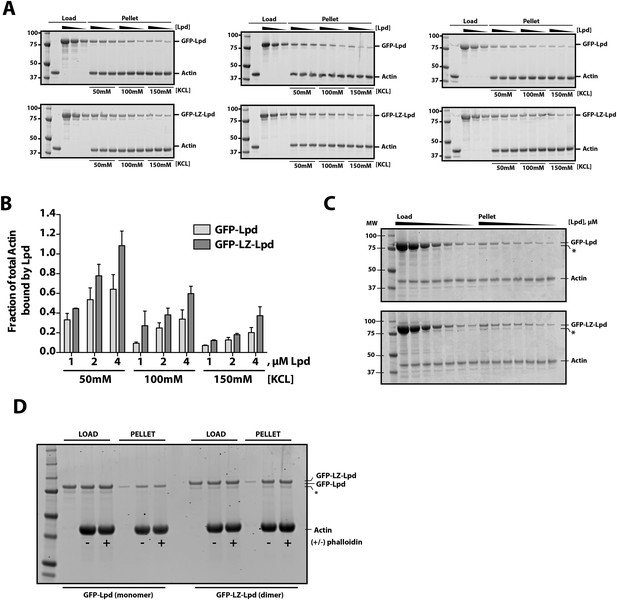
Interactions between filamentous actin, GFP-Lpd (850–1250aa), and GFP-LZ-Lpd (850–1250aa) measured by cosedimentation at different buffer ionic strengths.
(A) Monomeric GFP-Lpd850−1250aa and dimeric GFP-LZ-Lpd850−1250aa interact with filamentous actin in the presence of 50, 100, 150 mM KCl. SDS-PAGE from three experiments showing the cosedimentation of 2 µM filamentous actin (+4 µM dark phalloidin) in the presence of 1, 2, and 4 µM GFP-Lpd850−1250aa or GFP-LZ-Lpd850−1250aa (monomeric protein concentration). Buffer composition is 20 mM HEPES [pH 7], 50–150 mM KCl, 0.5 mM ATP, 0.5 mM MgCl2, 0.5 mM EGTA. (B) Average molar ratio of GFP-Lpd or GFP-LZ-Lpd bound to filamentous actin in the presence of 50, 100, and 150 mM KCl. Error bars represent S.D. of the mean (n = 3 experiments). (C) SDS-PAGE as in Figure 1H, showing the results of co-sedimentation of 1 µM filamentous actin in the presence of increasing concentrations of GFP-Lpd or GFP-LZ-Lpd (0–10 µM monomer concentration). (D) GFP-Lpd and GFP-LZ-Lpd interact with both ‘native’ and phalloidin stabilized actin filaments. Actin was polymerized at a concentration of 20 µM in the absence (termed ‘native’) or presence of an equal molar concentration of dark phalloidin (indicated by ‘+’). After 45 min, filamentous actin was combined with an equal volume of either 2 µM GFP-Lpd850−1250aa or GFP-LZ-Lpd850−1250aa and incubated for 1 hr before ultracentrifugation (also see ‘Materials and methods’). The final buffer composition was 20 mM HEPES [pH 7.0], 100 mM KCl, 1 mM TCEP, 0.5 mM ATP, 0.5 mM MgCl2, and 0.5 mM EGTA. (C, D) Asterisks (*) on SDS-PAGE gel marks partially translated or proteolyzed GFP-Lpd and GFP-LZ-Lpd that could not be removed during the purification.
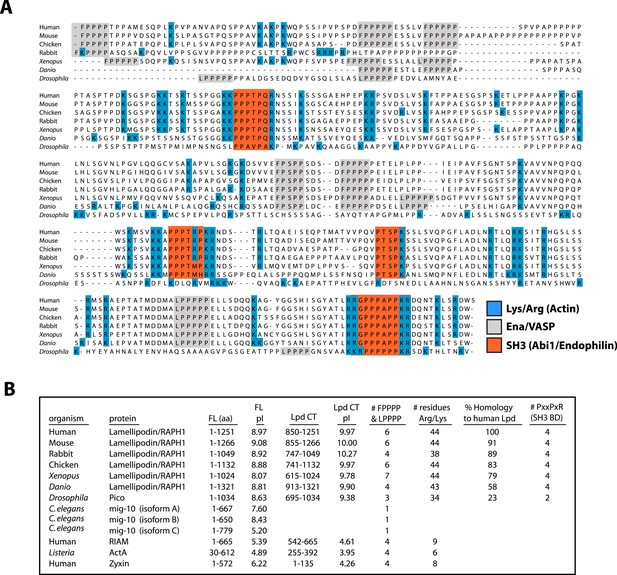
Conservation of Lpd (850–1250aa) amino acid sequence and isoelectric point (pI).
(A) Protein sequence alignment of human Lpd and homologs C-termini. Basic amino acid residues (arginine and lysine) are highlighted in blue. Gray boxes mark the location of the canonical Ena/VASP homology 1 (EVH1) BDs (i.e., FPPPP or LPPPP), while red boxes highlight the predicted Abi1/endophilin SH3 domain binding sites (i.e., PxxPxR). Secondary structure prediction algorithms suggest that the Lpd (850–1250aa) lacks secondary structure (data not shown). (B) Comparison of Lpd, Pico, mig-10, RIAM, ActA, and Zyxin pIs across the canonical Ena/VASP BD containing one or more FPPPP motifs. Domain boundaries for this region are termed, Lpd C-terminus (Lpd CT). The number of arginine and lysine residues were calculated across the region specified ‘Lpd C-terminus (Lpd CT)’. The number of Ena/VASP and SH3 domain binding sites were counted across the domain boundaries defined, Lpd CT, and are shown in columns five and six, respectively. The pIs for Lpd CT were calculated using EXPASY (Wilkins et al., 1999). Abbreviations are as follows: full-length (FL); binding-domain (BD); Lamellipodin (Lpd).
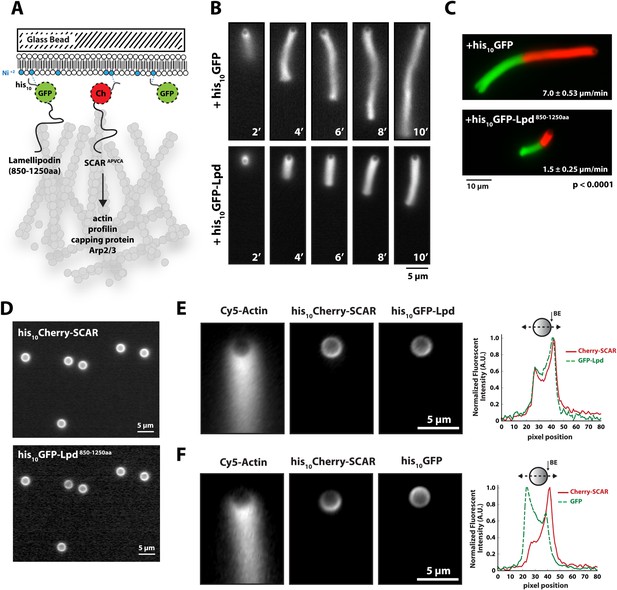
Membrane-tethered Lpd slows dendritic actin network assembly in vitro.
(A) Cartoon illustrating his10Cherry-SCARAPWCA, his10GFP-Lpd850−1250aa, and his10GFP tethered to a lipid coated beads (LCBs) containing DOGS-NTA(Ni) lipid (blue head groups). Actin network assembly on the bead surface is initiated by adding monomeric actin, profilin 1, Arp2/3, capping protein, and buffer containing KCl. (B, C) Membrane tethered his10-GFP-Lpd850−1250aa slows actin network assembly on LCBs. (B) Representative actin comet tails assembled in the presence of 7.5 µM actin (5% Alexa488-Actin), 3 µM hProfilin 1, 100 nM Arp2/3, 100 nM capping protein, and buffer containing 150 mM NaCl. LCBs (2.3 µm, 4% DOGS-NTA(Ni): 96% DOPC) were charged with 75 nM his10-Cherry-SCARAPWCA, plus 25 nM his10-GFP-Lpd850−1250aa or 25 nM his10-GFP (i.e., 75% his10-Cherry, 25% his10-GFP). Actin network assembly and disassembly was stopped at the indicated time points by combining the bead motility assay, 1:1, with 37.5 µM Latrunculin B-phalloidin mixture. Scale bar, 5 µm. (C) Representative actin comet tails assembled as in (B) for 5 min before transitioning from actin motility mix with 7.5 µM actin (5% Alexa488 labeled, GREEN) into an identical mix, but containing 7.5 µM actin (5% Cy3-Actin, RED). The length of Cy3-actin incorporated into the comet tail was measured to determine the growth velocity of multiple tails (n ≥ 50 tails). Error (±) represents the standard deviation of the mean (p-value = 3 × 10−29; two-tailed t-test for data sets with equal variance). Scale bar, 10 µm. (D) Homogenous distribution of his10-Cherry-SCARAPWCA and his10GFP-Lpd850−1250aa before initiating actin network assembly. Scale bar, 5 µm. (E, F) Spatial distribution of his10Cherry-SCARAPWCA, his10GFP-Lpd850−1250aa, and his10-GFP during steady state actin tail growth and recycling (30 min time point). Actin networks were assembled in the presence of 7.5 µM actin (5% Alexa488), 3 µM hProfilin 1, 100 nM Arp2/3, 100 nM Mm capping protein, and 3 µM hCofilin. (E) his10Cherry-SCARAPWCA and his10GFP-Lpd850−1250aa concentrate on the barbed end dense side of the actin comet tail. (F) his10-Cherry-SCARAPWCA concentrates on the barbed end dense side of the actin comet tail, while his10-GFP is excluded from the barbed end attachment zone. Line scans across LCBs are shown to the right. Scale bar, 5 µm.
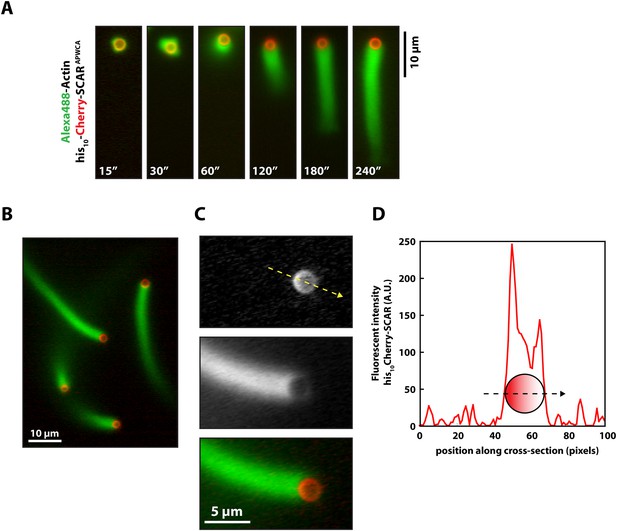
Actin based motility on lipid coated glass beads.
(A) Montage of actin comet tails frozen at different times point with 37.5 µM Latrunculin B-phalloidin containing buffer. Actin networks were assembled in the presence of 7.5 µM actin (5% Alexa488), 50 nM Arp2/3, 100 nM capping protein, 6 µM hPro1, 3 µM cofilin. Scale bar, 10 µm. (B) Image of actin comet tails at steady-state in the presence of cofilin dependent network recycling described in (A). Scale bar, 10 µm. (C) Images of asymmetric his10-Cherry-SCARAPWCA localization in the presence of actin comet tail. Scale bar, 5 µm. (D) Line scan across lipid coated bead surface in (C) showing fluorescent intensity of asymmetric his10-Cherry-SCARAPWCA.
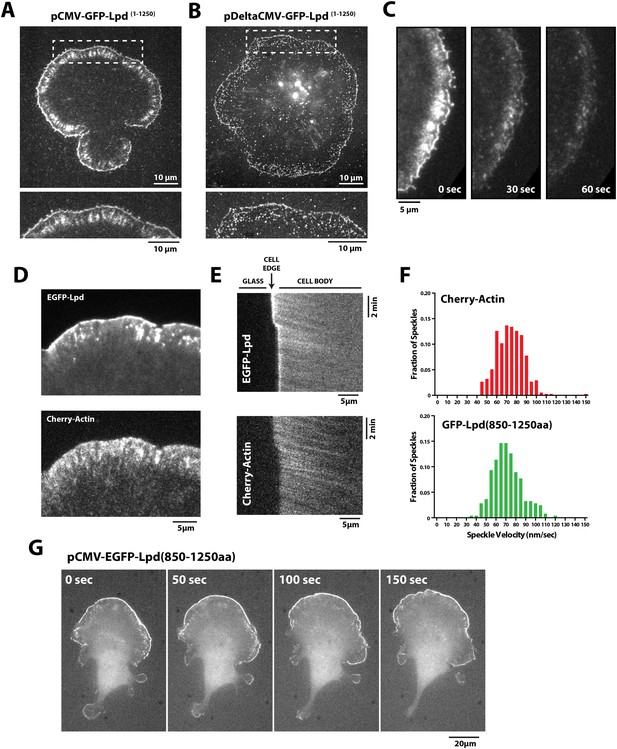
Lpd (850–1250aa) localizes to the leading edge membranes and undergoes retrograde flow with the actin cytoskeleton.
(A, B) Plasma membrane localization of GFP-Lpd1−1250aa visualized with TIRF microscopy in Xenopus Tissue Culture (XTC) cells spread on poly-L-lysine (PLL). GFP-Lpd1−1250aa was ectopically expressed from a (A) cytomegalovirus (CMV) or (B) DeltaCMV promoter. (B) Maximum intensity projection of a XTC cell expressing a single molecule concentration of GFP-Lpd1−1250aa. Scale bar, 10 µm. Leading edge membrane marked by dashed box is enlarged below. Scale bar, 5 µm. (C) Leading edge membrane localization of GFP-Lpd1−1250aa in XTC cell viewed by TIRF-M following the addition of 8 µM Jasplakinolide, 10 µM Latrunculin B, and 10 µM Y27632 (Rock kinase inhibitor) (Peng et al., 2011). Scale bar, 5 µm. (D) Representative image of XTC cell coexpressing GFP-Lpd850−1250aa and mCherry-Actin. Scale bar, 5 µm. (E) Kymographs show retrograde flow of GFP-Lpd850−1250aa and mCherry-Actin. Scale bar, 5 µm. (F) Histogram showing distribution of GFP-Lpd850−1250aa and mCherry-Actin speckle velocities. Mean speckle velocities of 71.9 ± 17.5 nm/s (n = 246 speckles) and 73.5 ± 14 nm/s (n = 373 speckles) were calculated for GFP-Lpd850−1250aa and mCherry-Actin, respectively. (G) GFP-Lpd850−1250aa localizes to the leading edge membrane of polarized mouse B16F1 cell migrating on laminin coated glass substrate. Scale bar, 20 µm.
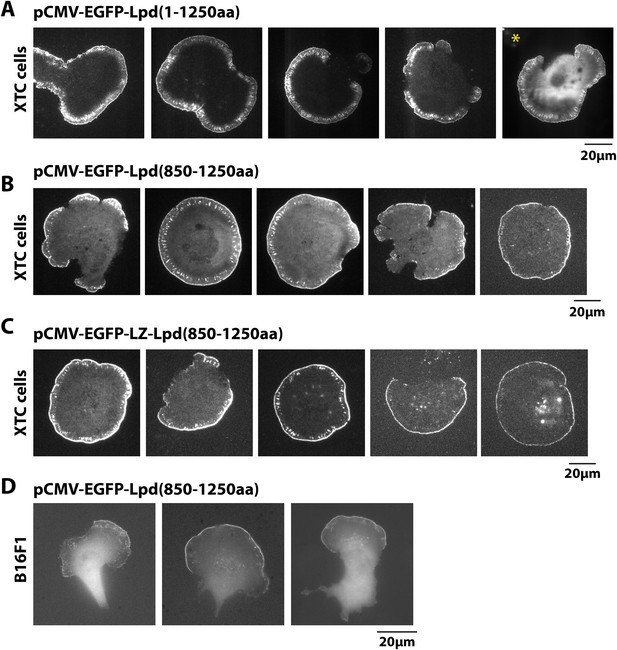
Localization of GFP-Lpd (850–1250aa) and GFP-LZ-Lpd (850–1250aa).
(A, C) Representative images showing the plasma membrane localization of (A) GFP-Lpd1−1250aa, (B) GFP-Lpd850−1250aa, and (C) dimeric GFP-LZ-Lpd850−1250aa visualized with TIRF microscopy in XTC cells spread on PLL. Image in (A), marked with asterisk (*), is a representative cell imaged using with wide-field epifluorescence. Note the cytoplasmic localization of GFP-Lpd1−1250aa is not visible by TIRF microscopy. Scale bar, 20 µm. (D) Representative images of GFP-Lpd850−1250aa localization in polarized B16F1 cell migrating on laminin coated glass and imaged with wide-field epifluorescence. Scale bar, 20 µm.
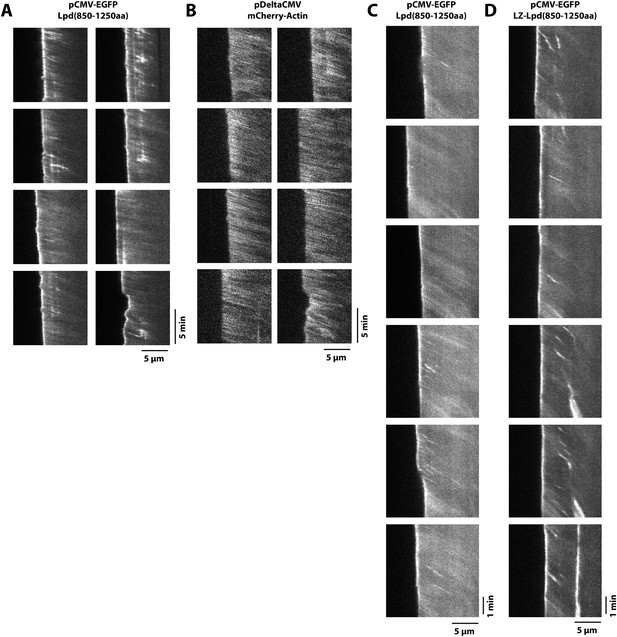
Retrograde flow of GFP-Lpd (850–1250aa) and GFP-LZ-Lpd (850–1250aa) with the actin cytoskeleton.
(A, B) Representative kymographs showing retrograde flow of (A) monomeric GFP-Lpd850−1250aa and (B) mCherry-Actin in XTC cells. Images were acquired every 5 s. Scale bars, 5 µm and 5 min. (C, D) Representative kymographs showing retrograde flow of (C) monomeric GFP-Lpd850−1250aa and (D) dimeric GFP-LZ-Lpd850−1250aa. Compared to (A, B), images were acquired every 2 s. Scale bars, 5 µm and 1 min.
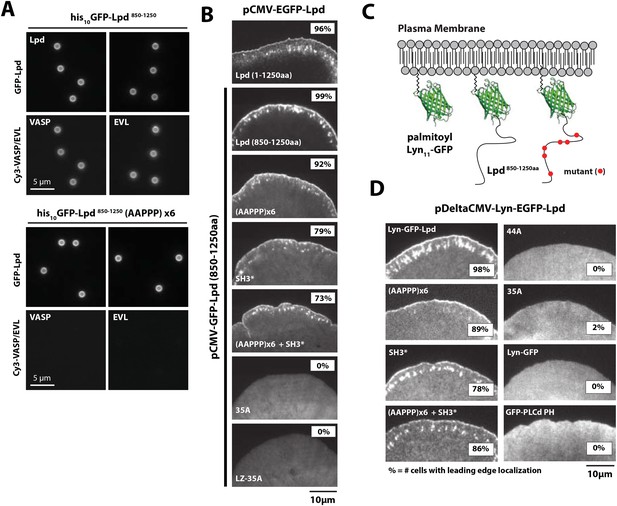
Interactions with Ena/VASP or Abi1/endophilin are not required for Lpd (850–1250aa) membrane localization.
(A) Lpd FPPPP peptide sequences are required to recruit Ena/VASP proteins to the lipid coated beads (LCBs). Glass microspheres were coated with SUVs containing DOPC/DOGS Ni-NTA lipids (96:4 molar ratio). LCBs were then incubated with 100 nM his10-GFP-Lpd850−1250aa, (wild-type and AAPPPx6 mutants) for 15 min, before being mixed with 500 nM Cy3-VASP or Cy3-EVL. Lpd mutant, (AAPPP)x6, cannot recruit purified Cy3-VASP or Cy3-EVL to LCBs. (B) Basic residues in flanking the Ena/VASP and Abi1/endophilin binding sites are required for leading edge localization of GFP-Lpd850−1250aa in XTC cells. Representative images of wild-type and mutant GFP-Lpd850−1250aa protein in XTC cells. Localization of full length GFP-Lpd1−1250aa (top panel) is shown for comparison. Scale bar, 5 µm. Refer to Figure 5—figure supplement 2 for amino acid sequences of each Lpd850−1250aa mutant. (C) Cartoon schematic showing palmitoylated Lyn-GFP and Lyn-GFP-Lpd (WT and FPPPP → AAPPPx6 mutant) anchored in the plasma membrane. The crystal structure of GFP was derived from Yang et al. (1996) (1GFL.pdb). (D) Constitutively membrane tethered Lyn-GFP-Lpd850−1250aa localizes to the leading edge. Leading edge localization of Lyn-GFP-Lpd850−1250aa does not require interactions with Ena/VASP proteins or Abi1/endophilin. Localization of Lyn-GFP and GFP-PLCδ (pleckstrin homology [PH] domain that binds to PI(4,5)P2), phenocopied the uniform membrane localization of Lyn-GFP-Lpd (35A and 44A). Scale bar, 10 µm. (B, D) The percentage of cells with leading edge localization is indicated in the upper right-hand corner of each representative image (n = 96–167 cells imaged for each GFP-Lpd850−1250aa construct expressed in XTC cells).
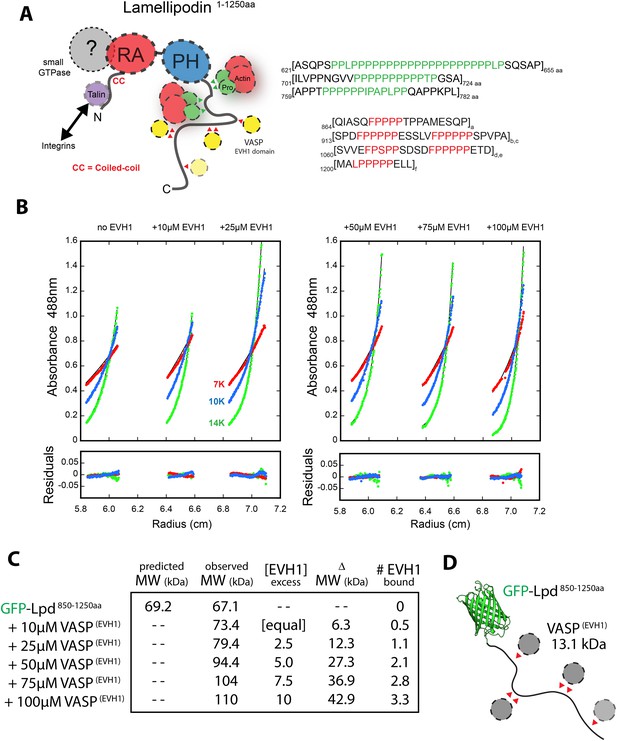
Lpd-VASP binding stoichiometry determined by sedimentation equilibrium.
(A) Cartoon showing domain organization of human Lpd (1–1250aa). (B) Analytical ultracentrifugation sedimentation equilibrium traces for GFP-Lpd850−1250aa in the absence (left) and presence of 10, 25, 50, 75, 100 µM VASP1−114aa EVH1 domain. (C) Table showing the predicted and observed molecular weight of GFP-Lpd850−1250aa in absence and presence of different VASP1−114aa EVH1 domain protein concentrations. (D) Cartoon showing GFP-Lpd850−1250aa with VASP1−114aa EVH1 domains (grey spheres) binding to FPPPP sites (red triangles). Based on the observed binding stoichiometry between GFP-Lpd850−1250aa and VASP1−114aa, we hypothesize that steric hindrance allows only a single EVH1 domain to interact with the tandem FPPPP motifs (i.e., SPDFPPPPPESSLVFPPPPPSPVPA and SVVEFPSPPSDSDFPPPPPETD). The crystal structure of GFP was derived from Yang et al. (1996) (1GFL.pdb).
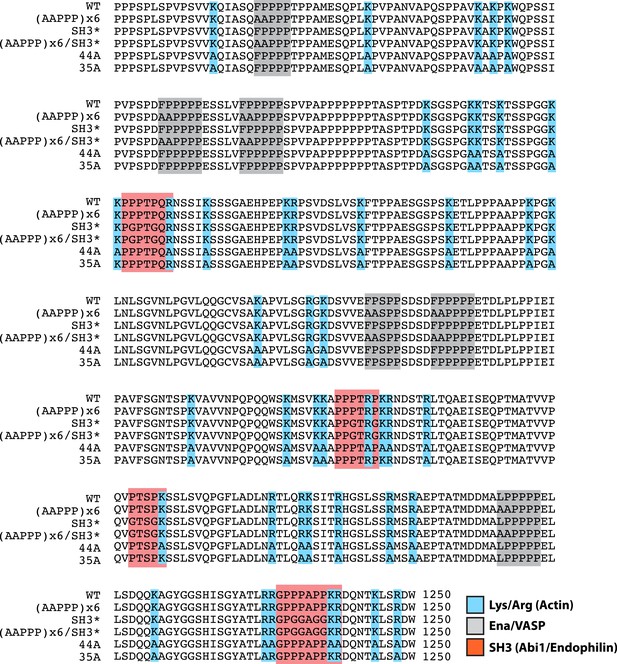
Lpd (850–1250aa) wild-type and mutant protein sequence alignment.
Protein sequence alignment of Lpd850−1250aa wild-type and mutants highlighting the separation of function mutations targeting either the actin BD (arg/lys; BLUE), Ena/VASP binding sites (GRAY), or Abi1/Endophilin SH3 domain binding sites (RED).
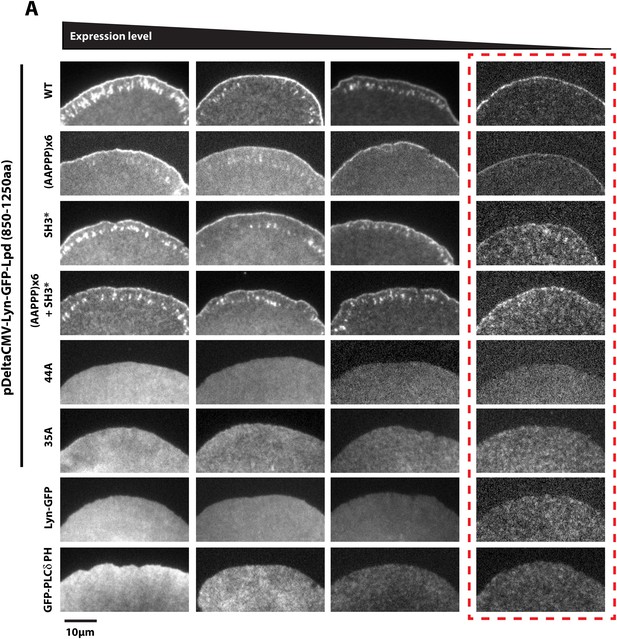
Membrane tethered Lyn-GFP-Lpd (850–1250aa) requires basic residue for leading edge localization.
(A) Plasma membrane localization of Lyn-GFP-Lpd850−1250aa, wild-type and mutants, visualized using TIRF microscopy. Mutations affecting the interaction with either Ena/VASP proteins (AAPPP)x6 or Abi1/Endophilin (SH3*) did not abolish the leading edge localization of Lyn-GFP-Lpd850−1250aa. Mutating all lysine and arginine residues (44A) or only those outside of the Abi1/Endophilin SH3 BDs (35A) eliminated the leading edge localization of Lyn- GFP-Lpd850−1250aa. Membrane localization of Lyn-GFP-Lpd (44A and 35A) phenocopy the uniform distribution of membrane anchored Lyn-GFP. Cell images highlighted with the red dashed box include representative images of cells expressing low level of Lyn- GFP-Lpd850−1250aa. The intensity of these images was scaled differently than images to the left to better visualize the localization.
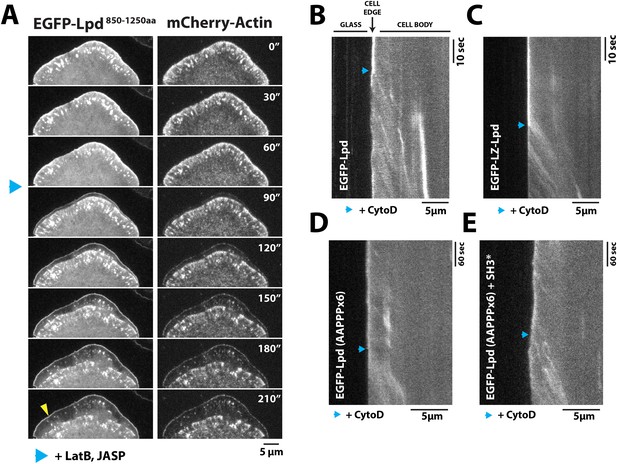
Dynamic actin filament assembly and free barbed ends are required for leading localization of GFP-Lpd (850–1250aa).
(A) Dynamic actin assembly is required for maintenance of GFP-Lpd850−1250aa leading edge localization. Image montage showing translocation of GFP-Lpd850−1250aa and mCherry-Actin toward the cell body, following the addition of 8 µM Jasplakinolide and 10 µM Latrunculin B. Note that a population of GFP-Lpd850−1250aa remains associated with the peripheral membrane after addition of Jasp-LatB (yellow arrowhead). Horizontal scale bar, 5 µm. Vertical scale bar, 2 min. (B–E) Barbed ends are required for plasma membrane localization of (B) GFP-Lpd850−1250aa, (C) GFP-LZ-Lpd850−1250aa, (D) GFP-Lpd850−1250aa (AAPPP)x6, and (E) GFP-Lpd850−1250aa (AAPPPx6 + SH3*). Representative kymographs showing membrane dissociation of GFP-Lpd850−1250aa, wild-type and mutants, following the addition of 100 nM Cytochalasin D (blue arrowhead). Horizontal scale bar, 5 µm.
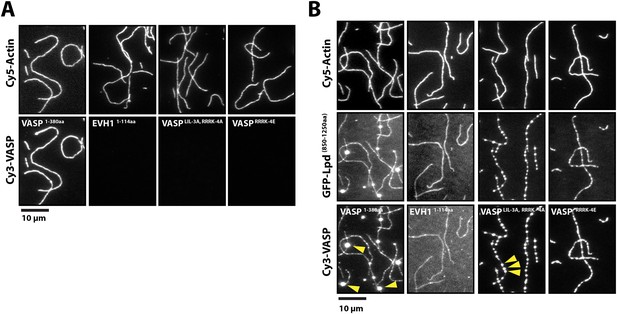
Lpd can simultaneously interact with VASP and filamentous actin.
(A) VASP EVH1 and FL VASP mutants cannot interact with actin filaments in vitro. Images highlight the inability of 200 nM (monomeric concentration) wild-type Cy3-VASP1−380aa, Cy3-VASP1−114aa (EVH1 domain), Cy3-VASPLIL-3A, RRRK-4A, and Cy3-VASPRRRK-4E to phalloidin stabilized actin filaments (20% Cy5 labeled). Buffer contains 20 mM HEPES [pH 7], 50 mM KCl, 1 mg/ml BSA, 1 mM TCEP. Scale bar, 10 µm. (B) GFP-Lpd850−1250aa can simultaneously interact with filamentous actin and VASP EVH1 domains. Colocalization of 500 nM monomeric GFP-Lpd850−1250aa phalloidin stabilized actin filaments (20% Cy5 labeled) in the presence of 200 nM (monomeric concentration) of wild-type Cy3-VASP1−380aa, Cy3-VASP1−114aa (EVH1 domain), Cy3-VASPLIL-3A, RRRK-4A (GAB and FAB mutant), or Cy3-VASPRRRK-4E (FAB mutant). Note the formation of large clusters containing VASP and Lpd (yellow arrowheads). Scale bar, 10 µm.
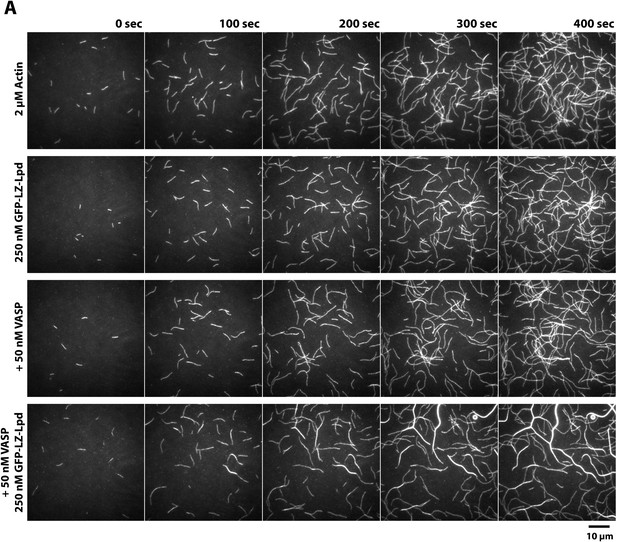
Lpd and VASP synergistically bundle actin filaments.
(A) Montage of single actin filaments polymerizing in the presence of 2 µM actin (20% Cy5 labeled) and TIRF buffer containing 100 mM KCl. Compared to actin filaments elongating in the presence of 50 nM VASP (tetrameric concentration) or 250 nM GFP-LZ-Lpd850−1250aa (dimer concentration). The combined presence of VASP and GFP-LZ-Lpd850−1250aa produces large actin bundles. Scale bar, 10 µm.
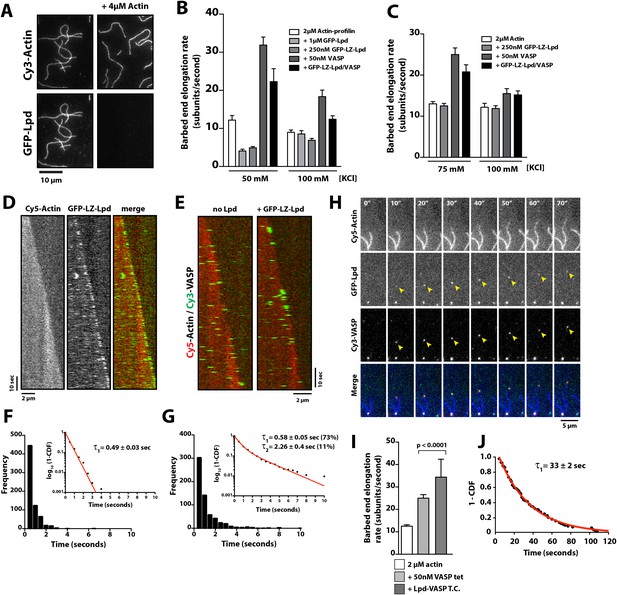
Lpd enhances VASP barbed end processivity.
(A) Monomeric actin antagonizes GFP-Lpd850−1250aa actin filament binding. Visualization of 500 nM GFP-Lpd850−1250aa in the absence or presence of 4 µM monomeric actin in the presence of buffer containing 20 mM HEPES [pH 7.0], 50 mM KCl, 1 mg/ml BSA, 1 mM TCEP, and 25 µM Latrunculin B. Scale bar, 10 µm. (B) GFP-Lpd850−1250aa (1 µM, monomer concentration) and GFP-LZ-Lpd850−1250aa (0.25 µM, dimer concentration) slow barbed end elongation in the presence of 2 µM profilin-Mg-ATP-actin (5% Cy5 labeled) and TIRF buffer containing 50 mM KCl. (C) Single actin filament elongation rates measured as in (B), but in the presence of 2 µM actin (20% Cy5) with TIRF buffer containing 75–100 mM KCl. (B, C) Error bars represent the standard deviation of the mean (n ≥ 30 barbed end elongation rates measured per condition). (D) Dimeric GFP-LZ-Lpd850−1250aa localizes to sides and barbed ends of elongating actin filaments. Kymographs showing the localization of 50 nM GFP-LZ-Lpd850−1250aa (green) to a single actin filament polymerized in the presence of 2 µM Mg-ATP-Actin (20% Cy5, red). Scale bars, 2 µm and 10 s. (E) Visualization of processive barbed end associated Cy3-VASP tetramers (green) in the absence or presence of 200 nM GFP-LZ-Lpd850−1250aa. Actin filaments were polymerized in the presence of 2 µM Mg-ATP-Actin (20% Cy5, red). Scale bar, 2 µm and 10 s. (F, G) Calculation of Cy3-VASP barbed end dwell times in the absence (F) or presence of 200 nM GFP-LZ-Lpd850−1250aa (G) decorated actin filaments. Histogram plots of Cy3-VASP barbed end associated dwell times with insets of the log10(1-cumulative distribution frequency) fit with a (F) single exponential curve for Cy3-VASP alone (τ1 = 0.49 ± 0.03 s, n = 673 molecules) or (G) Cy3-VASP in the presence of 200 nM GFP-LZ-Lpd850−1250aa (τ1 = 0.58 ± 0.05 s (73%, fast), τ2 = 2.3 ± 0.4 s (27%, slow), n = 632 molecules). Note that the dwell times for Cy3-VASP in (F) are shorter than previously reported (Hansen and Mullins, 2010). This due to Cy5-Actin being a less favorable substrate for barbed end incorporation compared to Alexa488-Actin. (H) Clustered his10-GFP-Lpd850−1250aa increases the processivity of Cy3-VASP. Image montage showing colocalization of the Cy3-VASP (5 nM) and his10-GFP-Lpd850−1250aa (50 nM) on actin filament barbed end elongating in the presence of 2 µM Actin (20% Cy5) and TIRF buffer contains 75 mM KCl. Note the intensity of the actin filament decreases when the VASP-Lpd complex is associated with the growing actin filament barbed end, indicating that unlabeled vs Cy5-labeled actin is more favorably incorporated. Scale bar, 5 µm. (I) Lpd-VASP barbed associated complexes incorporate actin monomers at a faster velocity, as compared to actin filament elongating in the presence of 50 nM tetrameric VASP. Error bars represent the standard deviation of the mean (p-value = 7 × 10−12; two-tailed t-test for data sets with unequal variance). (J) Calculation of the barbed end dwell times for Cy3-VASP and his10-GFP-Lpd850−1250aa complexes. Plot of 1-CDF was best fit to a single exponential curve, yielding τ1 = 33 ± 2 s (n = 87 complexes).
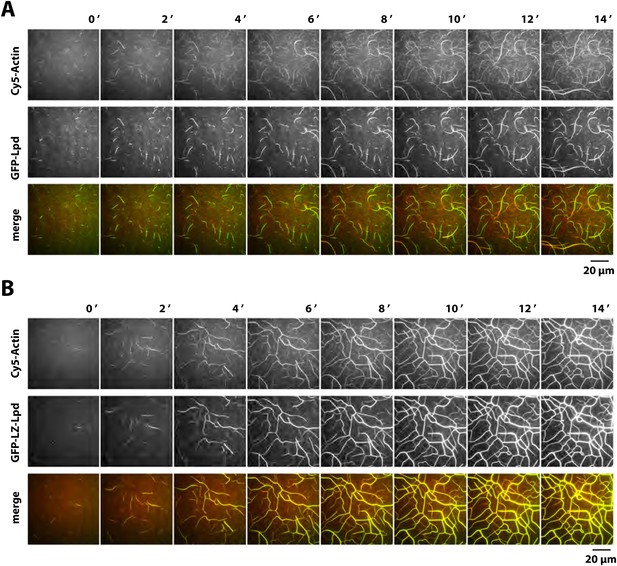
Lpd dependent actin filament bundling.
(A, B) Lpd bundles dynamically elongating actin filaments. Montage showing single actin filaments elongating and bundling in the presence of 2 µM actin (5% Cy5 labeled) and either (A) 1 µM GFP-Lpd850−1250aa or (B) 1 µM GFP-LZ-Lpd850−1250aa (dimer concentration equals 0.5 µM) in TIRF buffer containing 50 mM KCl. Note that the intensity of single actin filaments is faint due to the low percentage of Cy5-labeled actin and low laser intensity used to prevent camera pixel saturation by the bright actin filament bundles. Scale bar, 20 µm.
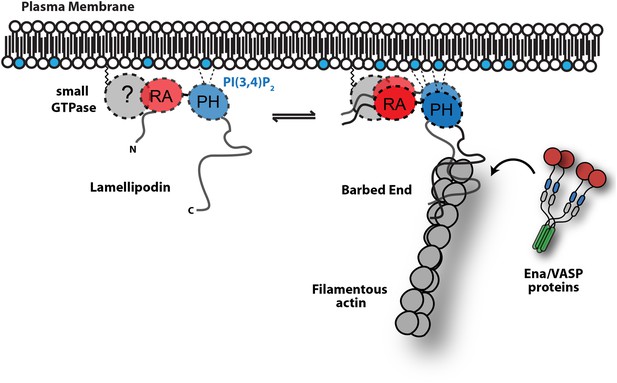
Model.
Based on the canonical model (Krause et al., 2004), Lpd is recruited to actin based membrane protrusions through interactions with phosphatidylinositol lipids (i.e., PI(3,4)P2) and possibly small GTPases (i.e., Ras or Rho family). Similar to the Grb protein family, Lpd is predicted to form homo-dimers mediated by interactions between the coiled-coil and tandem RA-PH domain. We find that the C-terminus of Lpd (residues 850–1250) is sufficient for recruiting Lpd to leading edge membrane where it directly interacts with free barbed ends and/or the sides of the actin filaments. Importantly, this interaction between Lamelliopodin and filamentous actin can occur independently to those mediated by Ena/VASP proteins or SH3 domains (i.e., Abi1/endophilin). However, Ena/VASP proteins recruited to actin based membrane protrusion can simultaneously associate with free actin filament barbed ends and Lpd. By this mechanism, we speculate that the lifetime of membrane targeted and barbed end associated Ena/VASP proteins are extended at the plasma membrane.
Videos
Localization of full length GFP-Lpd (1 −1250aa) expressed at single molecule concentrations from the DeltaCMV promoter in Xenopus Tissue Culture (XTC) cells.
Images were acquired with temporal resolution of 100 ms using Total Internal Reflection Fluorescence (TIRF)-M. Video plays at 15 frames per second. Scale bar, 5 µm.
Retrograde flow of GFP-Lpd850−1250aa and mCherry-Actin in XTC cells spread on poly-L-lysine (PLL) coated coverslips.
Cell was imaged with a temporal resolution of 5 s using TIRF-M at 23°C. GFP-Lpd850−1250aa and mCherry-Actin were ectopically expressed from either a CMV or DeltaCMV promoter, respectively. Video plays at 10 frames per second. Scale bar, 5 µm.
Leading edge membrane localization of GFP-Lpd850−1250aa in polarized B16F1 mouse melanoma cell migrating on laminin coated glass.
B16F1 cell was imaged with a temporal resolution of 10 s using wide-field epi fluorescence at 37°C. GFP-Lpd850−1250aa was ectopically expressed from a CMV promoter. Video plays at 10 frames per second. Scale bar, 20 µm.
Localization of membrane tethered Lyn-GFP-Lpd (850−1250aa) at the leading edge membrane of XTC cells spread on PLL coated coverslips expressed at single molecule concentrations from DeltaCMV promoter.
Images were acquired using TIRF microscopy with temporal resolution of 50 ms time. Video plays at 15 frames per second. Scale bar, 5 µm.
Localization of GFP-Lpd850−1250aa and mCherry-Actin in XTC cells following the addition of 8 µM Jasplakinolide and 10 µM Latrunculin B.
Images were acquired with a temporal resolution of 10 s using TIRF-M at 23°C. Drugs were added after 150 s of imaging. Video plays at 10 frames per second. Scale bar, 5 µm.
Localization of GFP-Lpd850−1250aa following the addition of 100 nM Cytochalasin D in a XTC cell imaged with a temporal resolution of 2 s using TIRF-M at 23°C.
Cytochalasin D was added at 214 s. Video plays at 50 frames per second. Scale bar, 5 µm.
Localization of GFP-LZ-Lpd850−1250aa following the addition of 100 nM Cytochalasin D in a XTC cell imaged with a temporal resolution of 2 s using TIRF-M at 23°C.
Cytochalasin D was added at 102 s. Video plays at 50 frames per second. Scale bar, 5 µm.
Dynamic actin filament association of 200 nM dimeric GFP-LZ-Lpd (850−1250aa) in the presence of 2 µM Mg-ATP (20% Cy5 labeled) visualized by TIRF-M.
For clarity, only GFP-LZ-Lpd (850−1250aa) is shown. Images were acquired with temporal resolution of 0.5 s using TIRF-M. Video plays at 50 frames per second. Scale bar, 5 µm.
Visualization of a processive VASP-Lpd tip complex bound to the barbed end of a single actin filament.
Elongation of single actin filaments were visualized in the presence of 2 µM Mg-ATP actin (20% Cy5 labeled), 5 nM Cy3-VASP, 50 nM his10-GFP-Lpd850−1250aa, and buffer containing 75 mM KCl. Images were acquired with temporal resolution of 2 s using TIRF-M. Video plays at 20 frames per second. Scale bar, 5 µm.
Additional files
-
Supplementary file 1
Table of plasmid DNA used for protein expression and cellular transfections.
- https://doi.org/10.7554/eLife.06585.030





