Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules
Figures
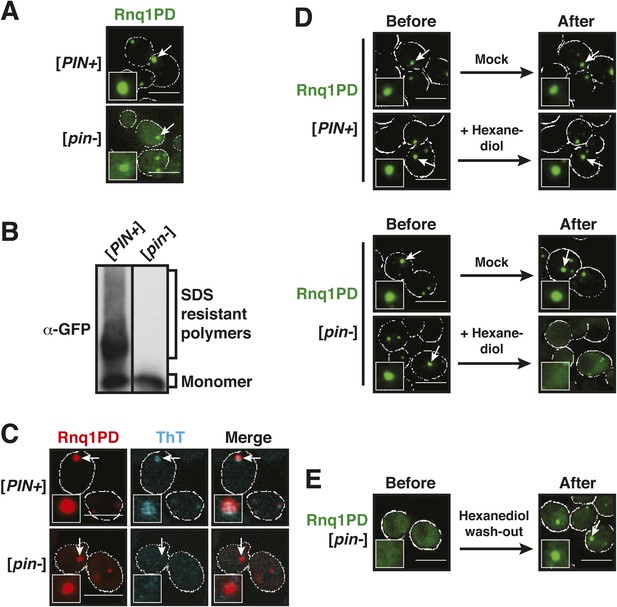
Prion-like domains can access two distinct aggregated states, only one of which is amyloid-like.
(A) Fluorescence microscopy of yeast cells expressing sfGFP-tagged Rnq1PD in [PIN+] and [pin−] cells. White lines indicate the cell boundaries. Scale bars: 5 µm. Also see related Figure 1—figure supplements 1–3. (B) Semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE) of [PIN+] and [pin−] cells containing a plasmid for expression of Rnq1PD-sfGFP. SDS-resistant amyloid polymers show slower migration in comparison to SDS-soluble monomers. Proteins were detected by immunoblotting with a GFP-specific antibody. (C) Thioflavin T (ThT) staining of [PIN+] and [pin−] cells expressing Rnq1PD-mCherry from a plasmid. Note that only amyloid-like assemblies can be stained with ThT. (D) 1,6-hexanediol treatment specifically disrupts non-amyloid Rnq1PD assemblies and not amyloids. Fluorescence time-lapse microscopy of [PIN+] (top panel) and [pin−] cells (bottom panel) expressing Rnq1PD-sfGFP. Time points are before treatment (Before) and 38 min after treatment with 10% 1,6-hexanediol (After). In the control condition (Mock) only media was added. Cells were permeabilized with 10 µg/ ml digitonin. See corresponding Video 1. (E) Cells expressing Rnq1PD-sfGFP were treated with 10% 1,6-hexanediol and digitonin for 1 hr to dissolve non-amyloid Rnq1PD assemblies (Before). Hexanediol was washed out and replaced with normal growth media (After), and the cells were observed with fluorescence microscopy. Also see corresponding Video 2.

Lsm4 has a prion-like C-terminal domain (underlined) that is enriched for asparagines (N) and glutamines (Q) and contains hydrophobic residues (L, V, I, M, F).
N and Q are highlighted in red and hydrophobic residues are highlighted in green.
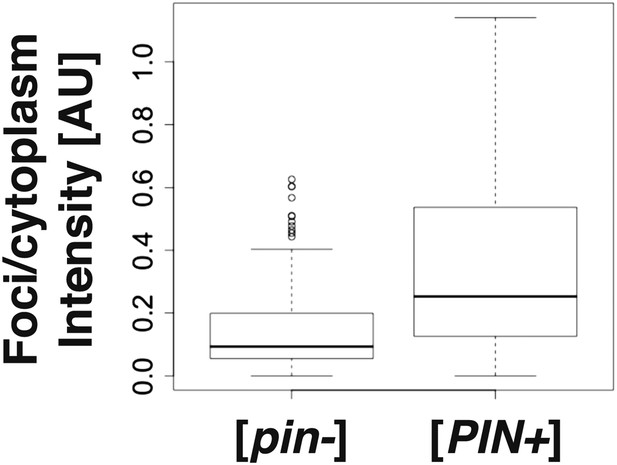
Rnq1PD shows a different aggregation pattern in [pin−] and [PIN+] cells.
The graph shows the foci-to-cytoplasm ratio of yeast cells expressing sfGFP-tagged Rnq1PD (number of cells analyzed from three independent experiments: [pin−] 95 cells, [PIN+] 91 cells). Using the unpaired Student's t-test, the values are significantly different with p = 5.5 × 10−08.

The amino acid sequence of the prion domain of Rnq1 resembles that of FG repeat-containing low-complexity domains of nucleoporins (hydrophobic aromatic residues in an asparagine- and glutamine-rich polar sequence background).
Aromatic hydrophobic residues are highlighted in green and N and Q residues are highlighted in red.
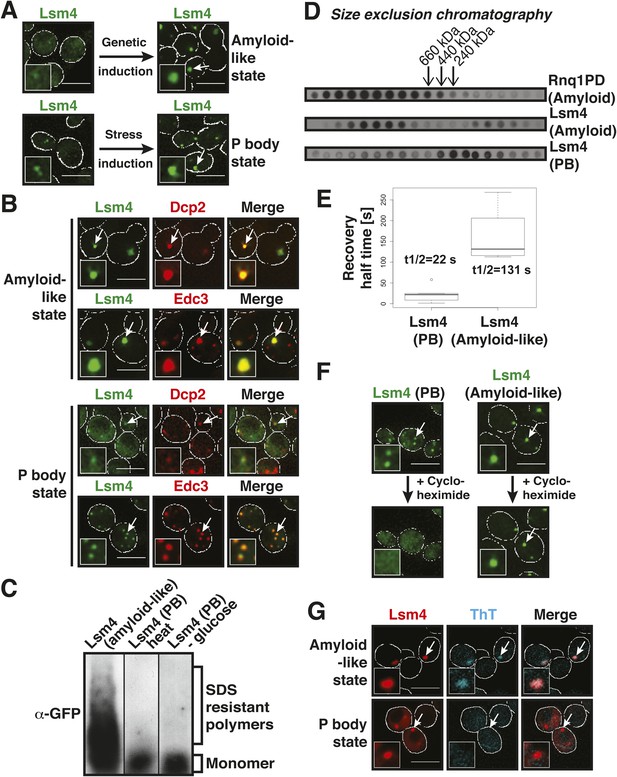
The prion-like protein Lsm4 does not adopt an amyloid-like conformation in P bodies.
(A) Genetically induced and stress-induced Lsm4 assemblies are morphologically similar. Endogenous GFP-tagged Lsm4 in the amyloid-like state and PB state was investigated by fluorescence microscopy. PB formation was induced by glucose starvation for 1 hr (Stress induction). Genetic induction of Lsm4-GFP assembly into an amyloid-like state was through overexpression of unlabeled Lsm4. White lines indicate the cell boundaries. Scale bars: 5 µm. (B) Fluorescence microscopy of Lsm4-GFP cells co-expressing the mCherry-tagged PB proteins Dcp2 or Edc3. Lsm4 assembly was induced genetically (amyloid-like state) or through heat stress at 46˚C for 10 min (P-body state). Note that both types of assemblies show co-localization with Dcp2 and Edc3. (C) Lsm4 in P bodies does not adopt an amyloid-like conformation. Comparative SDD-AGE analysis of Lsm4-GFP in the amyloid-like and PB state. P bodies were induced through heat stress or glucose depletion. (D) The Lsm4 RNP complex in P bodies has a different size than the amyloid-like Lsm4 complex. Size exclusion chromatography of Lsm4-GFP in the amyloid-like state and PB state. Cells expressing Rnq1PD from a plasmid were used as a control for amyloid. Molecular size standards were: thyroglobulin (660 kDa), ferritin (440 kDa), and catalase (240 kDa). Cells were stressed in glucose-deficient medium to induce PBs. (E) Fluorescence recovery after photobleaching analysis of Lsm4 in the P body state (induced by glucose depletion) or amyloid-like state. Dendra2-tagged Lsm4 was expressed from a plasmid. The median half-recovery times were 22 s for the PB state (n = 6) and 131 s for the amyloid-like state (n = 4). Using the unpaired Student's t-test, the values are significantly different with p = 0.0018. Also see related Figure 2—figure supplement 1. (F) Lsm4-containing P bodies are RNA-dependent, whereas amyloid-like Lsm4 assemblies are not. Cells containing Lsm4-GFP in the PB state (induced by glucose depletion) or amyloid-like state were treated with 100 µg/ml cycloheximide and observed by fluorescence time-lapse microscopy after 14 min. See corresponding Video 3. (G) P bodies are not stainable with the amyloid-specific dye ThT. Fluorescence microscopy of ThT-stained cells expressing mCherry-tagged Lsm4. Lsm4 assembly was induced genetically (amyloid-like state) or through glucose depletion (P-body state). Also see related Figure 2—figure supplement 2.
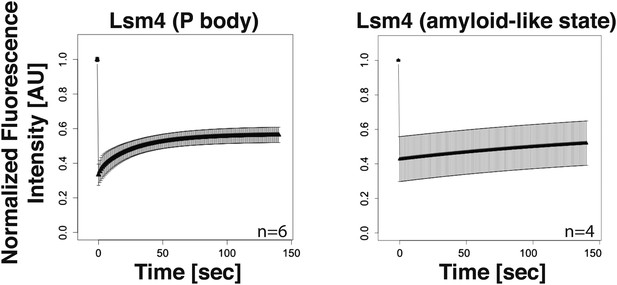
Lsm4 in P bodies shows faster turnover than Lsm4 in amyloid-like assemblies.
FRAP analysis was performed using the photoconvertible tag Dendra2. The recovery of the green fluorescence was followed over time after photoconversion. P-body formation was induced by glucose depletion. The normalized fluorescence intensity is shown. Error bars represent SEM.
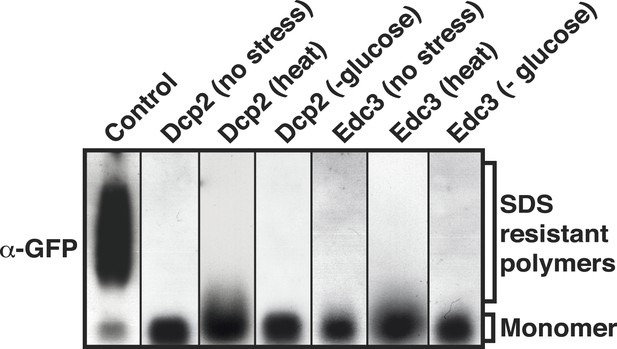
P-body proteins do not enter into an amyloid-like state upon stress.
SDD-AGE analysis of whole cell lysates from yeast cells that expressed GFP-tagged Dcp2 or Edc3 from their endogenous loci. P-body formation was induced through heat stress (46°C) or glucose depletion. The prion-like protein Nrp1-GFP in the amyloid-like state was used as a control (left).
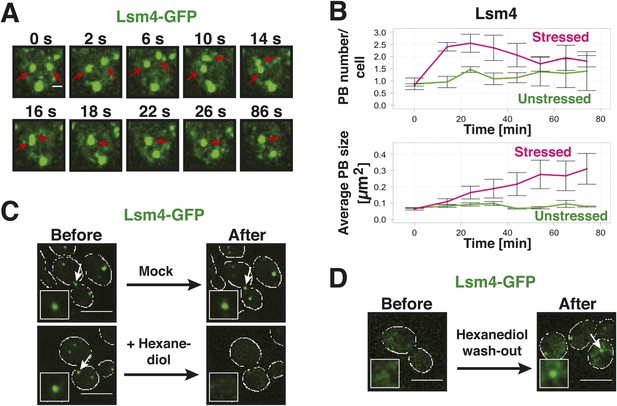
Yeast P-bodies are liquid droplets and not aggregates.
(A) P bodies behave as liquid-like droplets. Fluorescence time-lapse microscopy of stressed cells expressing Lsm4-GFP from the endogenous locus. Time points are indicated in seconds above. Two fusing PBs are indicated by red arrows. White scale bar: 2 µm. (B) Quantification of the PB fusion behavior over time using GFP-tagged Lsm4 as a marker. At time point 0, the medium was changed to synthetic medium lacking glucose (red curves). Control cells received complete synthetic medium (green curves). Values given are PB number/cell and the average PB size [µm2]. At last 150 cells were analyzed. Error bars represent SEM. (C) P bodies are sensitive to 1,6-hexanediol, suggesting that they are liquid-like. Cells expressing Lsm4-GFP from the endogenous promoter were stressed in medium without glucose for 30 min (Before). 5% 1,6-hexanediol or medium (Mock) was added and the cells were analyzed after 30 min (After). Scale bars: 5 µm. Also see corresponding Video 4 and related Figure 3—figure supplement 1. (D) The hexanediol effect is reversible. Stressed cells expressing Lsm4-GFP were treated with 5% 1,6-hexanediol for 1 hr (Before). Hexanediol was washed out with fresh medium and an image was acquired after 12 min (After). Also see corresponding Video 5.
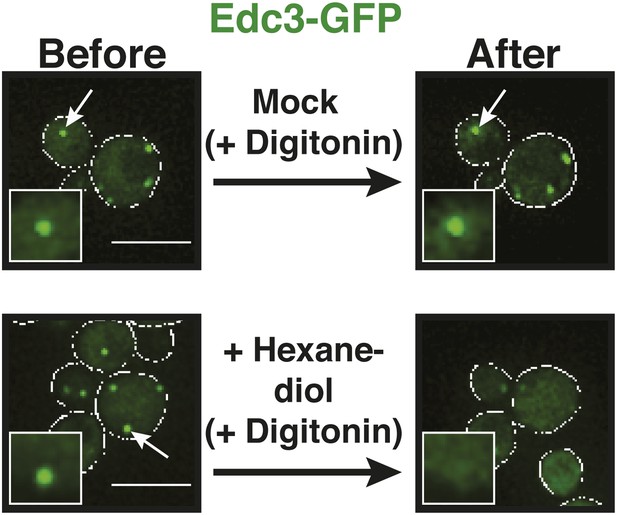
The aliphatic alcohol 1,6-hexanediol causes rapid disassembly of P bodies.
Fluorescence microscopy of the P-body marker Edc3-GFP expressed from the endogenous locus. Cells were stressed in medium without glucose for 15 min and 10 µg/ml digitonin with or without 10% 1,6-hexanediol was added. Images were taken before hexanediol treatment (Before) and 2 min after treatment (After). White lines indicate the cell boundaries. Scale bars: 5 µm.
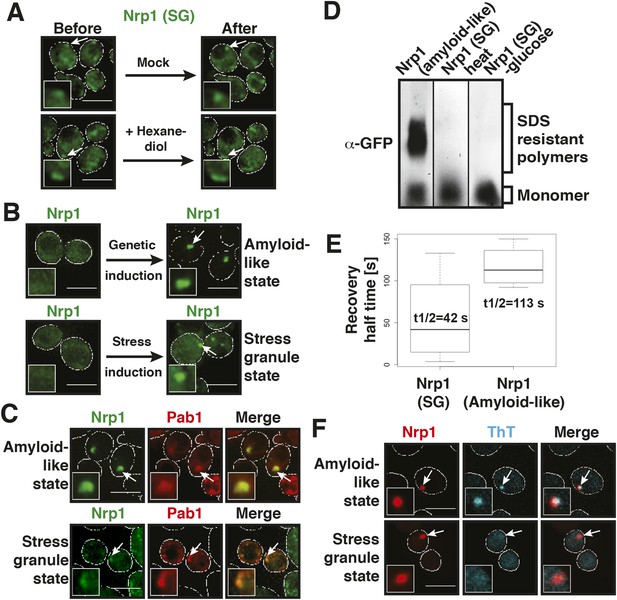
Stress granules have properties of solid aggregates, but do not depend on amyloid-like conversions for their formation.
(A) 1,6-hexanediol does not disrupt stress granules. Cells expressing Nrp1-GFP from the endogenous promoter as a marker for stress granules were stressed for 15 min in glucose-deficient medium (Before). An image was taken 6 min after the addition of 10% hexanediol (After). The cells were treated with 10 µg/ml digitonin to make them more permeable to hexanediol. The control (Mock) received only medium +10 µg/ml digitonin. Note that P bodies dissolve within less than 2 min under the same conditions (see Figure 3—figure supplement 1 and Figure 4—figure supplement 1). Also see corresponding Video 6. (B) The amyloid-like and stress granule states of Nrp1 are morphologically similar. Fluorescence microscopy of cells expressing GFP-tagged Nrp1 from the endogenous promoter. Nrp1 assembly was induced genetically (amyloid-like state) or through 1 hr glucose starvation (stress granule state). White lines indicate the cell boundaries. Scale bars: 5 µm. (C) Genetically induced and stress-induced Nrp1 assemblies are morphologically similar and recruit Pab1. Fluorescence microscopy analysis of Nrp1-GFP cells expressing mCherry-tagged Pab1. Stress was induced through heat shock at 46°C 10 min. (D) Stress does not induce an amyloid-like state in Nrp1. SDD-AGE of lysates from cells expressing Nrp1-Cerulean from the endogenous promoter. Nrp1 assembly was induced genetically (amyloid-like state) or through stress (stress granule state, SG). Also see related Figure 4—figure supplement 4. (E) Analysis of Nrp1 turnover in the stress granule state (induced by glucose depletion) and amyloid-like state. Nrp1-Dendra2 was expressed from a plasmid. The recovery half times of Nrp1 in the SG state (median = 42 s, n = 4) and in the amyloid-like state (median = 113 s, n = 4) are shown. p value = 0.0927 (unpaired Student's t-test). Also see related Figure 4—figure supplement 3. (F) Stress granules are not stainable with ThT. Cells expressing mCherry-tagged Nrp1 were treated with ThT. Assembly of Nrp1 was induced genetically (amyloid-like state) or through glucose depletion (stress granule state). Also see related Figure 4—figure supplement 2.
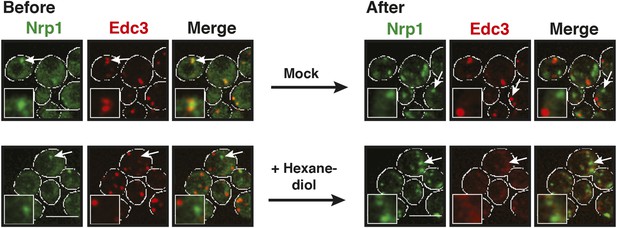
P bodies but not stress granules are sensitive to hexanediol.
Cells expressing Nrp1-GFP and Edc3-mCherry from the endogenous loci were stressed in medium without glucose for 80 min (Before). 5% 1,6-hexanediol or medium (Mock) was added and the cells were analyzed by fluorescence microscopy after 2 hr (After). White lines indicate the cell boundaries. Scale bars: 5 µm.
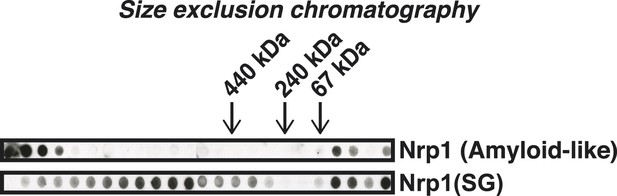
Stress granule-associated and amyloid-like Nrp1 form different complexes in yeast cell lysate.
Size-exclusion chromatography of lysates from cells expressing Nrp1-Cerulean from the endogenous promoter. Nrp1 in the amyloid-like state and SG state (glucose starvation) is compared. Molecular weight standards were: ferritin (440 kDa), catalase (240 kDa), and BSA (bovine serum albumin) (67 kDa).
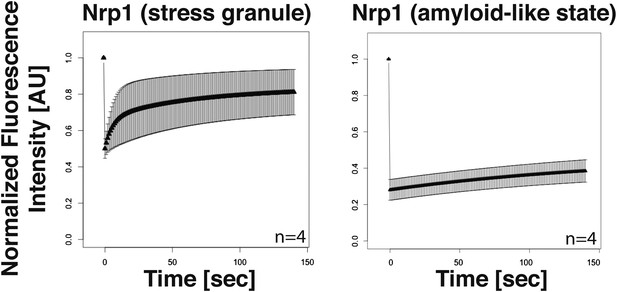
Analysis of the turnover of Nrp1 in the stress granule and the amyloid-like state.
Dendra2-tagged Nrp1 was used for FRAP analysis. The recovery of the green fluorescence was followed over time after photoconversion. The SG state was induced through glucose depletion. The normalized fluorescence intensity in the SG state (left panel) and amyloid-like state (right panel) are compared. Error bars represent SEM.
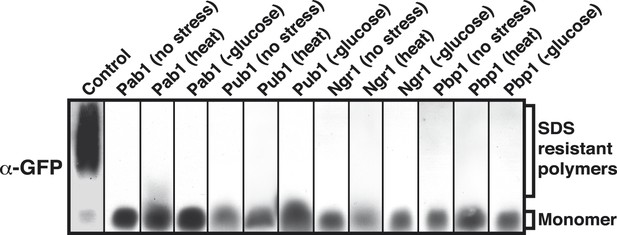
Prion-like stress granule proteins do not transition into an amyloid-like state.
SDD-AGE analysis of endogenous GFP-tagged Pab1, Pub1, Ngr1 or Pbp1 in unstressed and stressed yeast cells. Stress granules were induced through heat stress (46°C) or glucose depletion. Nrp1-GFP in the amyloid conformation was used as a control (left).
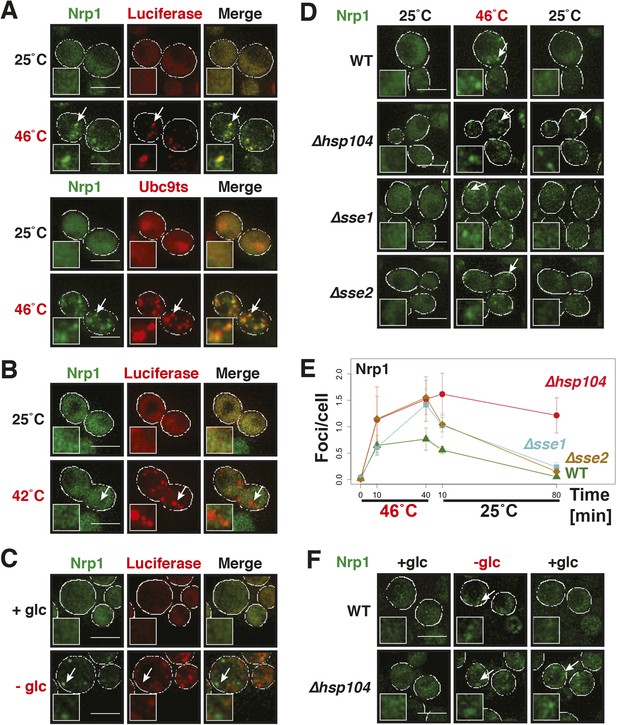
Yeast stress granules are functional protein aggregates, which are dissolved by disaggregases.
(A) The stress granule protein Nrp1 is co-deposited with misfolding-prone proteins during robust heat shock. Fluorescence microscopy of yeast cells expressing Nrp1-GFP from the endogenous locus and mCherry-tagged misfolding-prone proteins (mutated luciferase and Ubc9ts) from a plasmid. The cells were exposed to a 10-min heat shock at 46°C. White lines indicate the cell boundaries. Scale bars: 5 µm. Also see related Figure 5—figure supplements 1–3 and Videos 7–9. (B) Nrp1 is less aggregation prone than luciferase. Same as (A), except that the cells were exposed to a mild heat shock at 42°C for 10 min. Note that under these conditions only luciferase forms aggregates. Also see related Figure 5—figure supplement 4. (C) Nrp1 and luciferase coalesce into distinct structures under starvation conditions. Same as (A), except that the cells were stressed by glucose starvation for 1 hr (-glc). See corresponding Video 10. Also see related Figure 5—figure supplement 5. (D) Dissolution of stress granules is dependent on disaggregases. Fluorescence time-lapse microscopy of yeast cells expressing GFP-tagged Nrp1 from the endogenous locus. Wild-type cells are compared to strains with genetic deficiencies (Δhsp104, Δsse1, or Δsse2). Shown are images after 40 min at 46˚C and during recovery after 80 min at 25˚C. Also see Video 11. (E) Quantification of the number of foci/cell in the strains shown in (D). At least 290 cells were analyzed for each strain from three independent experiments. Error bars are SEM. (F) Hsp104 is also required for the disassembly of stress granules induced by glucose starvation. Wild-type or Δhsp104 cells expressing Nrp1-GFP from the endogenous locus were exposed to glucose starvation for 1 hr (image taken after 40 min at -glc) and observed 80 min after glucose-containing growth medium was added.
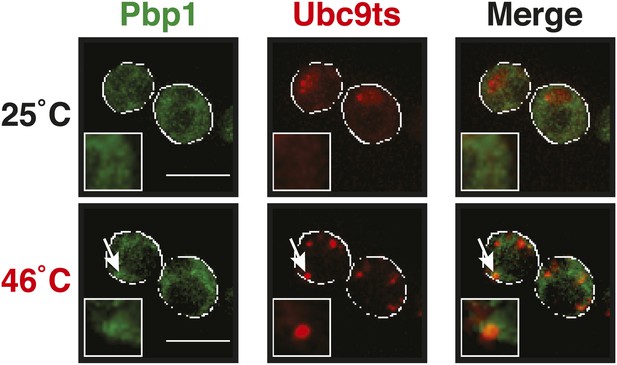
The stress granule protein Pbp1 is co-deposited with the misfolding-prone protein Ubc9ts during robust heat shock.
Fluorescence microscopy of yeast cells expressing Pbp1-GFP from the endogenous locus and mCherry-tagged Ubc9ts from a plasmid. Cells were exposed to a heat shock at 46°C. White lines indicate the cell boundaries. Scale bars: 5 µm.
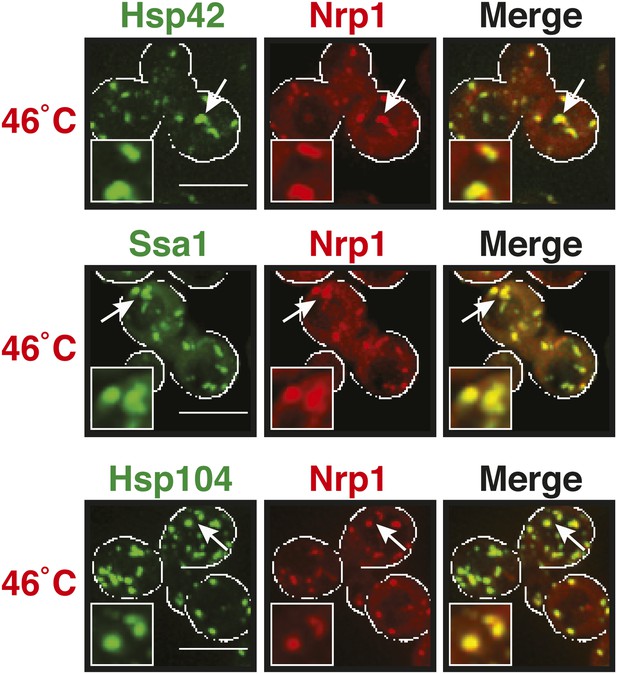
The stress granule protein Nrp1 co-localizes with chaperones during robust heat shock.
Fluorescence microscopy of yeast cells expressing Nrp1-mCherry from a plasmid and GFP-tagged Hsp42, Ssa1, or Hsp104 from the endogenous locus. Cells were exposed to a heat shock at 46°C. White lines indicate the cell boundaries. Scale bars: 5 µm.
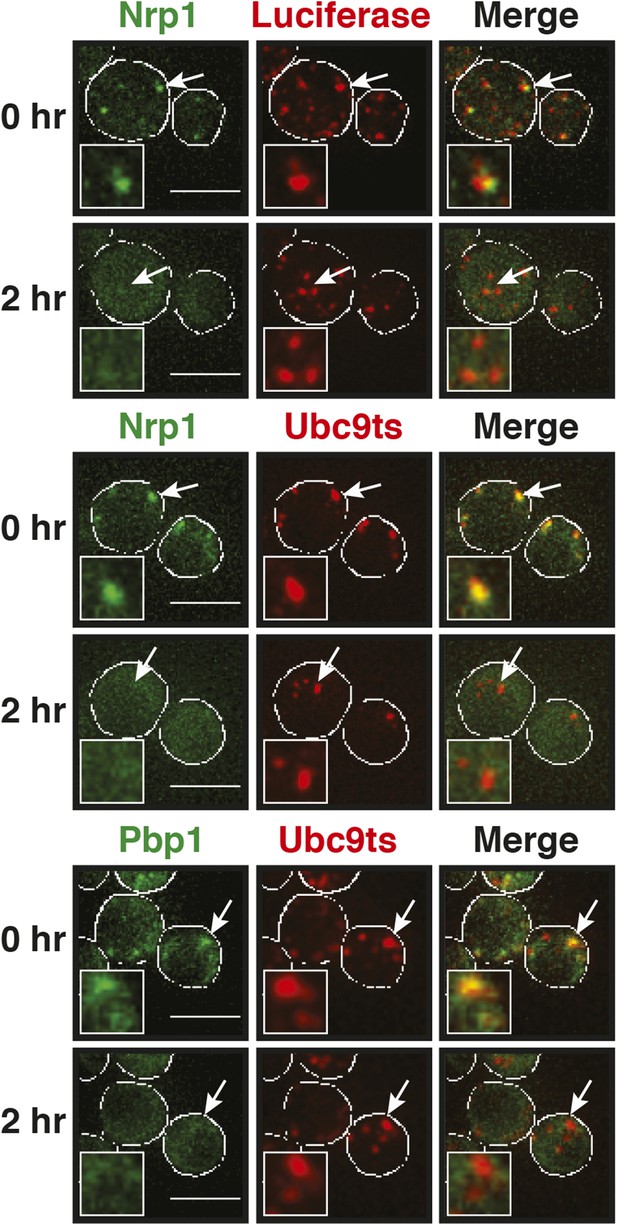
Stress granules dissolve faster than protein aggregates.
Yeast cells expressing Nrp1-GFP or Pbp1-GFP from the endogenous locus were transformed with plasmids for the expression of mCherry-tagged misfolding-prone proteins (mutated luciferase or Ubc9ts). Cells were exposed to a robust heat shock at 46°C for 2 hr (time point: 0 hr) and then transferred back to 25°C and observed after 2 hr (time point: 2 hr). Also see the corresponding Videos 7–9. White lines indicate the cell boundaries. Scale bars: 5 µm.
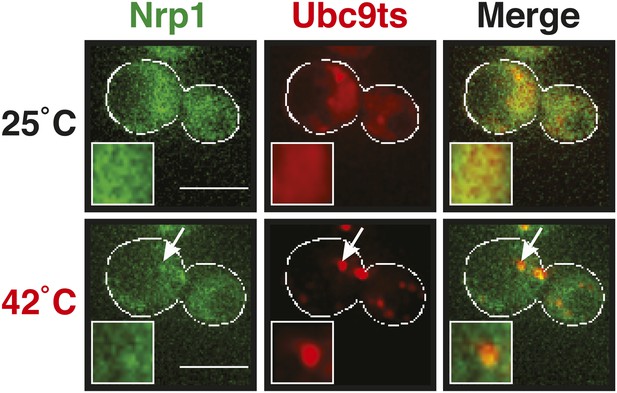
A mild heat shock does not lead to co-deposition of stress granule components and misfolding-prone proteins.
Fluorescence microscopy of yeast cells expressing Nrp1-GFP from the endogenous locus and mCherry-tagged Ubc9ts from a plasmid. The cells were exposed to a mild heat shock at 42°C. White lines indicate the cell boundaries. Scale bars: 5 µm.
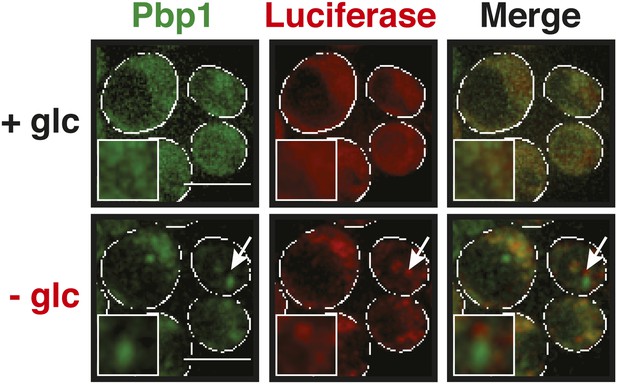
Stress granules and misfolding-prone proteins do not form mixed aggregates under glucose depletion conditions.
Yeast cells expressing Pbp1-GFP from the endogenous locus and mutant mCherry-tagged luciferase from a plasmid were exposed to glucose starvation stress (−glc). White lines indicate the cell boundaries. Scale bars: 5 µm.
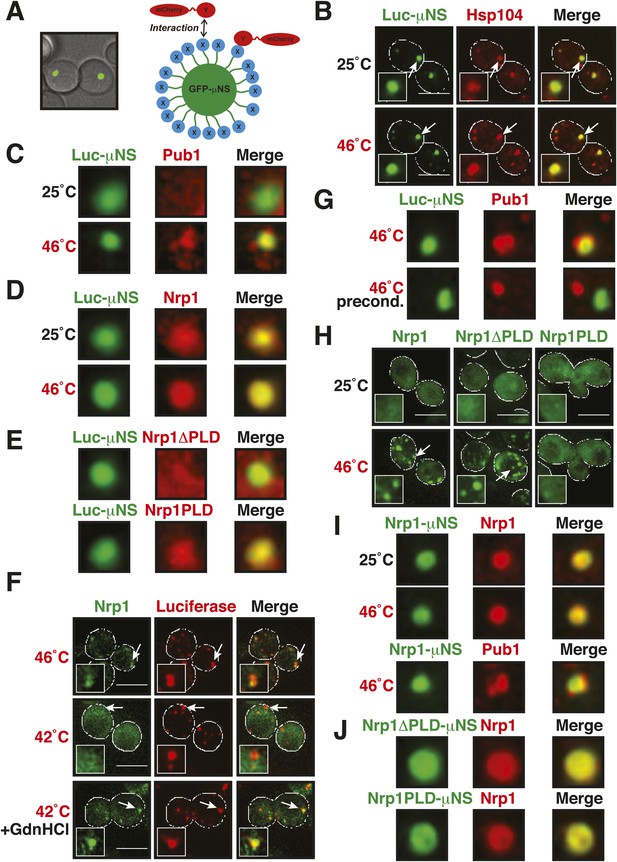
Stress granule assembly is redundant and highly adaptable.
(A) Using a genetically encoded particle to study dynamic interactions in living yeast cells. Left: a fragment of the viral capsid protein μNS comprising the 250 C-terminal amino acids forms self-assembling particles in yeast cells. The particles were visualized using an N-terminal sfGFP tag. Right: interaction assay. Fusion of a protein X to sfGFP-tagged μNS particles allows interaction studies with a mCherry-tagged protein Y. (B) μNS particles carrying mutant luciferase on the surface interact with endogenous mCherry-tagged Hsp104. Cells were observed before and after a 10-min heat shock at 46°C. White lines indicate the cell boundaries. Scale bars: 5 µm. Also see related Figure 6—figure supplements 1, 2. (C) Same as (B), except that cells were used in which mCherry-tagged Pub1 was expressed from the endogenous locus. Only one representative μNS particle is shown at high magnification. Note that Pub1 only interacts with luciferase in cells exposed to robust heat stress. Also see related Figure 6—figure supplements 3–5. (D) Same as (C), except that mCherry-tagged Nrp1 was mildly overexpressed from a plasmid carrying an ADH1 promoter. Note that Nrp1 interacts with luciferase already in unstressed cells, and that the amount of Nrp1 accumulating on the particle is strongly increased upon heat stress. (E) Same as (D), except that the prion-like domain (PLD) of Nrp1 (Nrp1PLD) or a deletion mutant lacking the PLD (Nrp1ΔPLD) was observed at 25˚C. (F) The cellular chaperone machinery prevents interactions between misfolded proteins and stress granule components. Cells expressing Nrp1-GFP from the endogenous locus and mCherry-tagged mutated luciferase from a plasmid were exposed to a 10-min heat shock at 42°C or 46°C. The cells in the bottom panel were exposed to 3 mM guanidinium hydrochloride (GdnHCl) to inhibit Hsp104. Also see related Figure 6—figure supplement 6. (G) Same as (C), except that the temperature was increased slowly from 25°C to 46°C (preconditioning). Note that preconditioning prevents co-assembly of stress granules and misfolded proteins. Also see related Figure 6—figure supplement 7. (H) PLDs mediate interactions only when present in high local concentrations. Yeast cells were transformed with plasmids for the expression of GFP-tagged wild-type Nrp1 or deletion mutants lacking the RNA-binding domain (RBD) (Nrp1PLD) or PLD domain (Nrp1ΔPLD). The resulting cells were exposed to heat shock. (I) Upon heat shock, stress granules form on µNS particles that present Nrp1 on the surface. Same conditions as (C) and (D). Also see related Figure 6—figure supplements 8–9. (J) Same as (I), except that mutants lacking the RBD (Nrp1PLD) or PLD domain (Nrp1ΔPLD) were presented on the particle. Note that both mutants are able to recruit full-length Nrp1 at 25˚C. Also see related Figure 6—figure supplement 10–12.
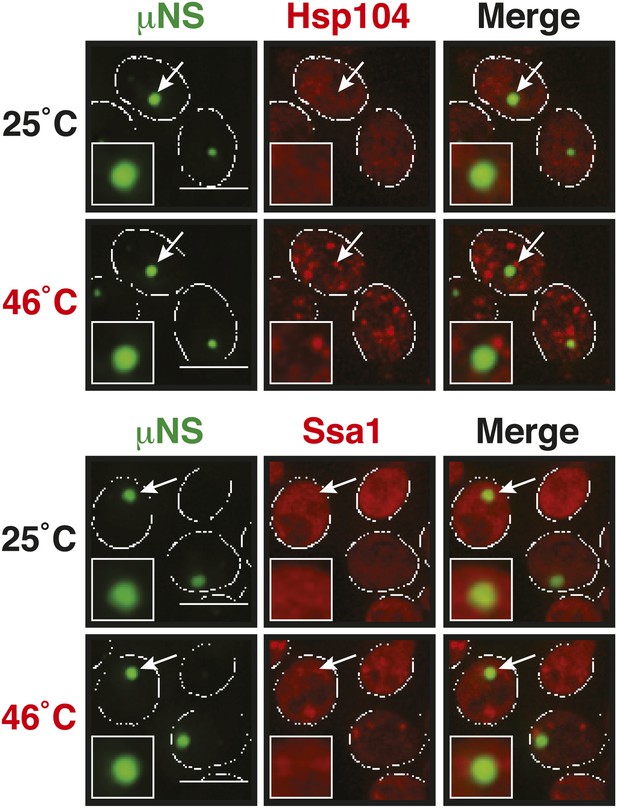
µNS particles do not interact with Hsp104 or Ssa1.
Cells expressing tdimer2-tagged Hsp104 and Ssa1 from the endogenous locus were observed before and after heat shock at 46°C. White lines indicate the cell boundaries. Scale bars: 5 µm.
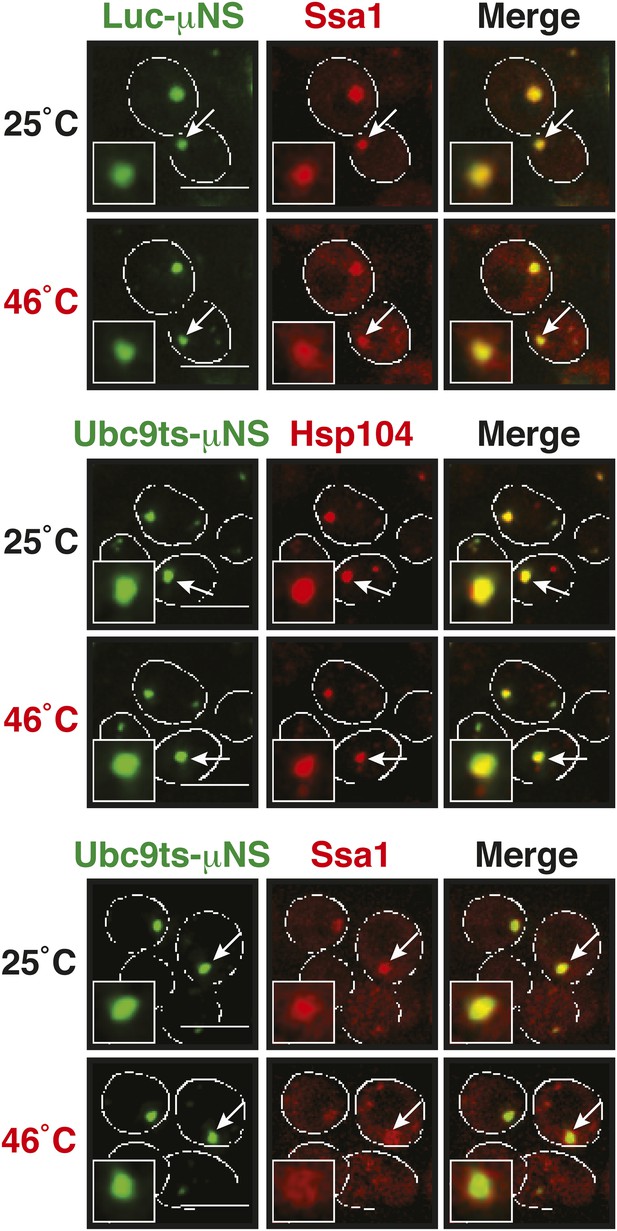
sfGFP-µNS particles carrying mutant luciferase or Ubc9ts on the surface interact with Hsp104 and Ssa1.
Cells expressing tdimer2-tagged Hsp104 and Ssa1 from the endogenous locus were observed before and after heat shock at 46°C. Note that Ubc9ts is already interacting with chaperones in unstressed cells, suggesting that a fraction of Ubc9ts is already misfolded under normal conditions. White lines indicate the cell boundaries. Scale bars: 5 µm.
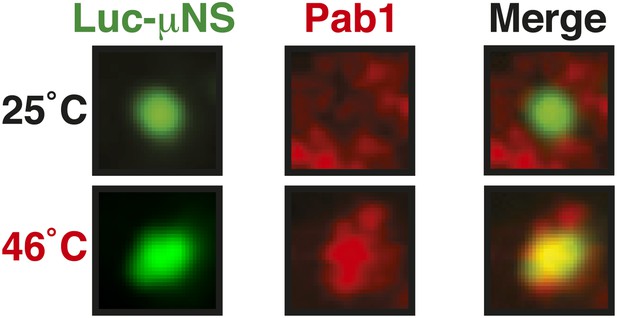
Misfolded proteins can nucleate stress granule formation under robust heat shock conditions.
Same as Figure 6—figure supplement 2, except that cells were used in which mCherry-tagged Pab1 was expressed from the endogenous locus. Note that Pab1 only interacts with luciferase in cells that are exposed to robust heat stress. For presentation purposes, only one representative μNS particles is shown at high magnification for each experiment.
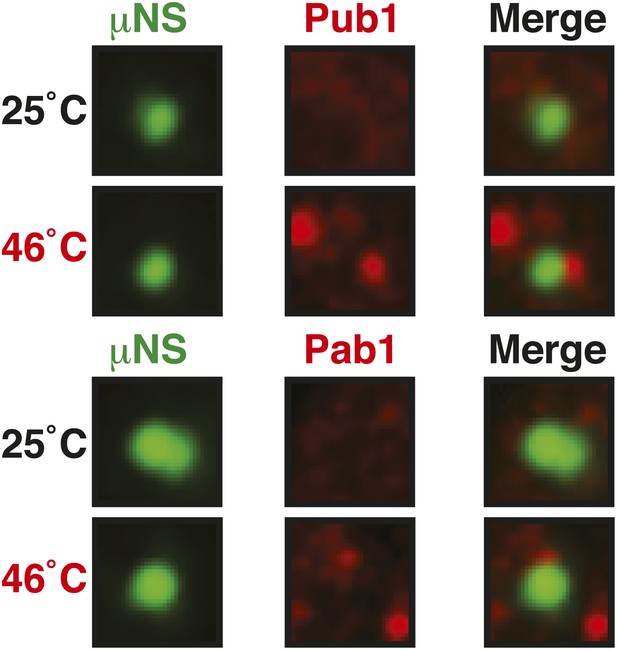
Control experiment showing that µNS particles do not interact with Pub1 or Pab1.
Same as Figure 6—figure supplement 1, except that strains were used that expressed mCherry-tagged Pub1 or Pab1 from the endogenous locus. For presentation purposes, only one representative μNS particles is shown at high magnification for each experiment.
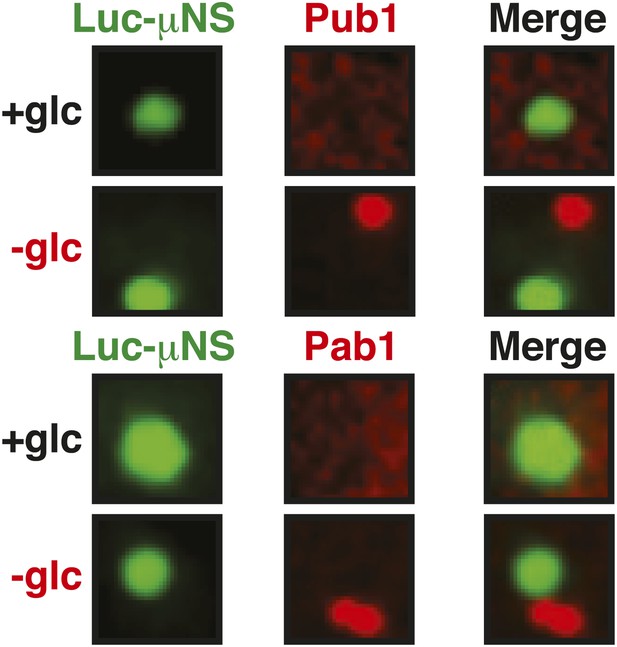
Misfolded proteins do not nucleate stress granule formation in glucose-deprived cells.
Same as Figure 6—figure supplement 2, except that strains were used that expressed mCherry-tagged Pub1 or Pab1 from the endogenous locus, and stress was induced by removing glucose from the medium for 1 hr. For presentation purposes, only one representative μNS particles is shown at high magnification for each experiment.
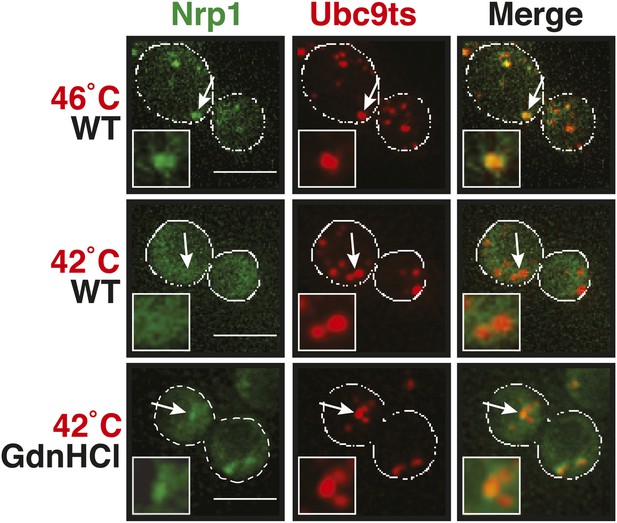
Chemical inhibition of Hsp104 leads to co-aggregation of misfolded proteins and stress granule components even under mild heat shock conditions.
Yeast cells expressing Nrp1-GFP from the endogenous locus and mCherry-tagged mutated Ubc9ts from a plasmid were exposed to heat shock at 42°C or 46°C. The cells in the bottom panel were exposed to 3 mM GdnHCl to inhibit Hsp104. White lines indicate the cell boundaries. Scale bars: 5 µm.
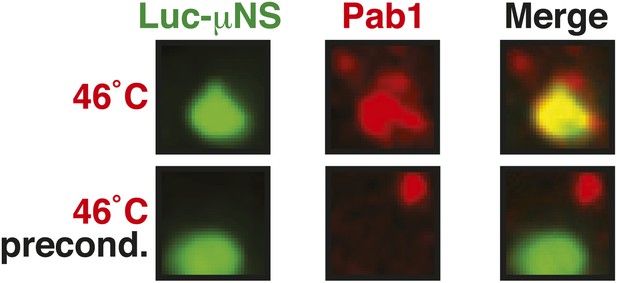
Misfolded proteins and stress granule components do not co-aggregate in preconditioned cells.
Same as Figure 6—figure supplement 3, except that the temperature was increased slowly from 25°C to 46°C (preconditioning). For presentation purposes, only one representative μNS particles is shown at high magnification for each experiment.
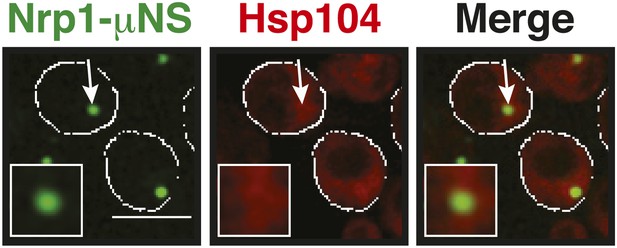
Control experiment showing that Nrp1-µNS particles do not interact with Hsp104.
A strain expressing tdimer2-tagged Hsp104 from the endogenous promoter was transformed with a construct for the expression of Nrp1-sfGFP-μNS. Note that particle-presented Nrp1 is not recognized by Hsp104 at 25˚C, indicating that it is not misfolded on the particle surface. White lines indicate the cell boundaries. Scale bar: 5 µm.
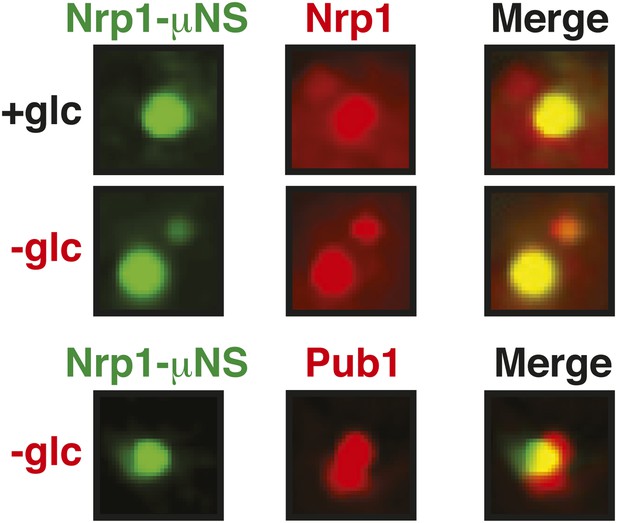
Nrp1 can nucleate stress granules upon glucose starvation stress.
Same as Figure 6—figure supplement 8, except that strains were used that expressed mCherry-tagged Nrp1 and Pub1 and cells were exposed to glucose starvation for 1 hr. For presentation purposes, only one representative μNS particles is shown at high magnification for each experiment.
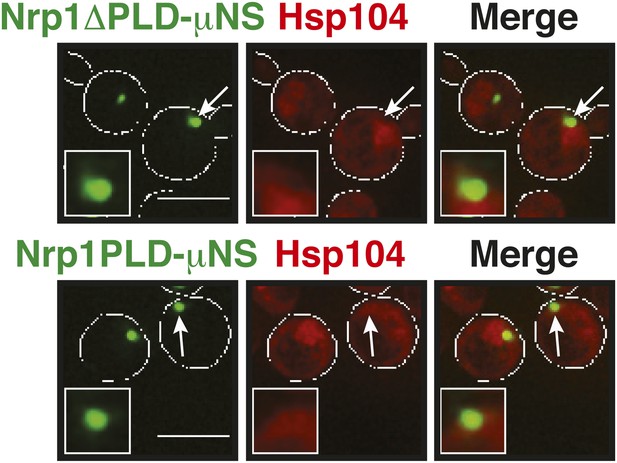
Same as Figure 6—figure supplement 8 except that mutants lacking the RBD (Nrp1PLD) or PLD domain (Nrp1ΔPLD) were presented on the particle.
Note that both deletion mutants are not recognized by Hsp104 at 25˚C, suggesting that they are folded and accessible on the particle surface. White lines indicate the cell boundaries. Scale bars: 5 µm.
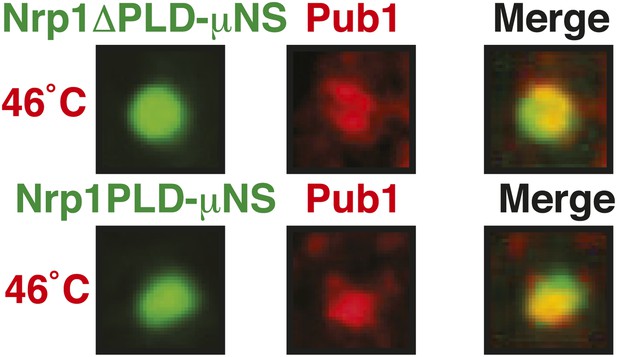
The PLD as well as the RBD of Nrp1 can nucleate stress granule formation.
Same as Figure 6—figure supplement 10, except that a strain was used in which mCherry-tagged Pub1 was expressed from the endogenous locus and the cells were exposed to a heat shock at 46°C. For presentation purposes, only one representative μNS particles is shown at high magnification for each experiment.
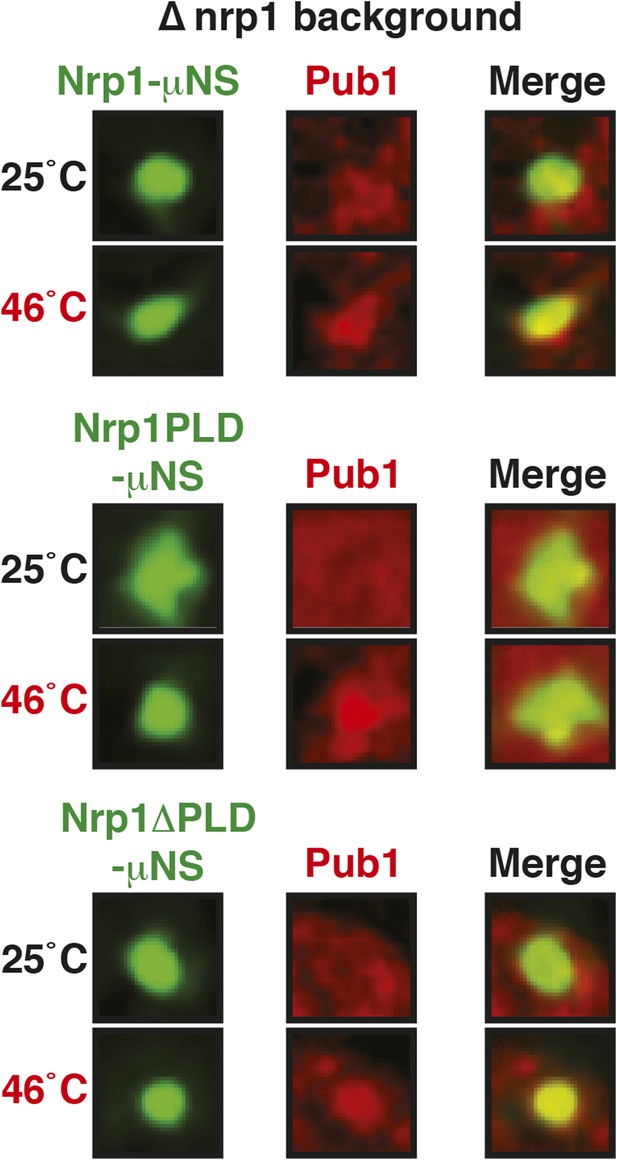
The PLD or RBD of Nrp1 can nucleate stress granule formation through heterotypic interactions.
A Pub1-mCherry-expressing strain lacking Nrp1 (Δnrp1) was transformed with a construct for expression of GFP-µNS fused to full-length Nrp1, Nrp1PLD, or Nrp1RBD. The cells were exposed to a heat shock at 46°C. For presentation purposes, only one representative µNS particle is shown at high magnification.
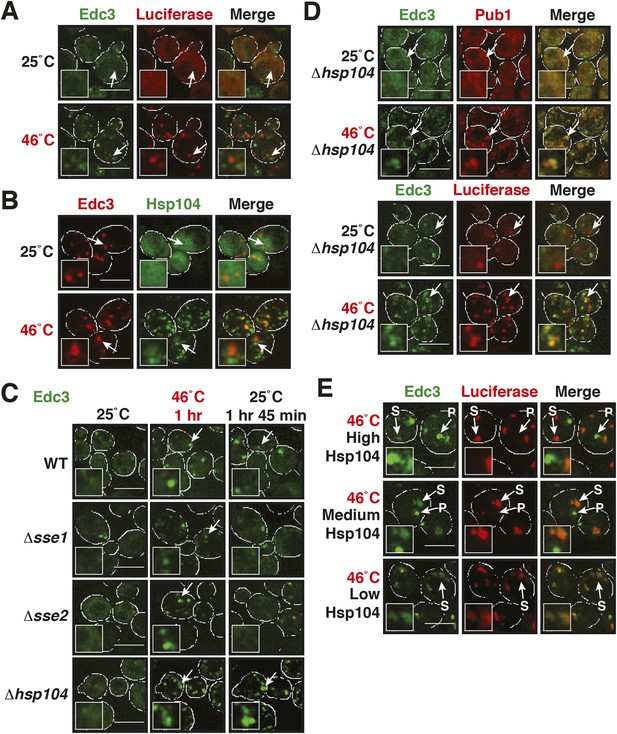
Maintenance of yeast P body integrity requires Hsp104.
(A) The P-body protein Edc3 shows only minor co-localization with misfolded proteins during robust heat stress. Fluorescence microscopy of endogenous GFP-tagged Edc3 and plasmid-expressed mCherry-tagged mutant luciferase. The cells were stressed at 46°C for 10 min. Cell boundaries are indicated in white. Scale bars: 5 µm. Also see related Figure 7—figure supplement 1. (B) Edc3 shows only limited spatial overlap with Hsp104. Fluorescence microscopy of plasmid expressed mCherry-tagged Edc3 and Hsp104-GFP expressed from the endogenous locus. The cells were subjected to heat stress. Also see related Figure 7—figure supplement 2. (C) Edc3-positive assemblies show different morphologies and behavior in the absence of disaggregases. Fluorescence time-lapse microscopy of yeast cells expressing GFP-tagged Edc3 from the endogenous locus. Wild-type cells are compared to strains lacking disaggregases (Δhsp104, Δsse1, or Δsse2) after 1 hr heat stress at 46˚C and 1 hr 45 min after recovery at 25˚C. Also see Video 12 and related Figure 7—figure supplement 3. (D) P-body components co-assemble with stress granules in heat-stressed cells in the absence of Hsp104. Yeast cells expressing endogenous Edc3-GFP and endogenous Pub1-mCherry or aggregation-prone mCherry-tagged luciferase from a plasmid in Hsp104-deficient cells were subjected to 1 hr heat stress at 46˚C. (E) The amount of stress granule-localized Edc3 (denoted with S) decreases with increasing amounts of Hsp104. P denotes P bodies. Yeast cells were used that expressed Edc3 under the endogenous promotor and mCherry-tagged mutated luciferase from a plasmid. Endogenous hsp104 was deleted and substituted with plasmid-expressed Hsp104 under control of a GPD - glyceraldehyde-3-phosphate dehydrogenase (high), ADH1 - alcohol dehydrogenase 1 (medium), or SUP35 (low) promoter. Cells were observed after 1 hr at 46˚C. Also see Video 13.
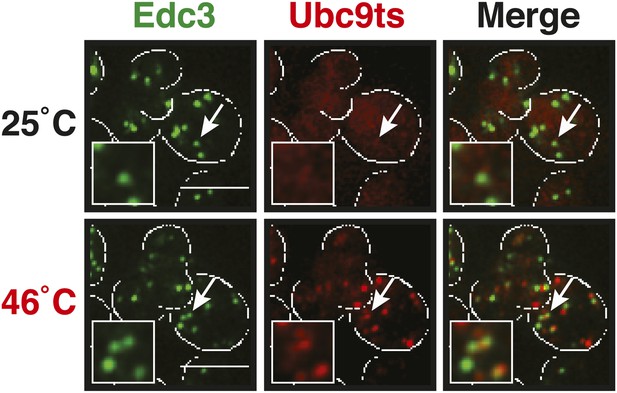
P bodies do not co-aggregate with misfolded proteins under robust heat shock conditions.
Edc3-GFP was expressed from the endogenous locus and mCherry-tagged Ubc9ts from a plasmid. Cells were observed before and after heat shock at 46°C. White lines indicate the cell boundaries. Scale bars: 5 µm.
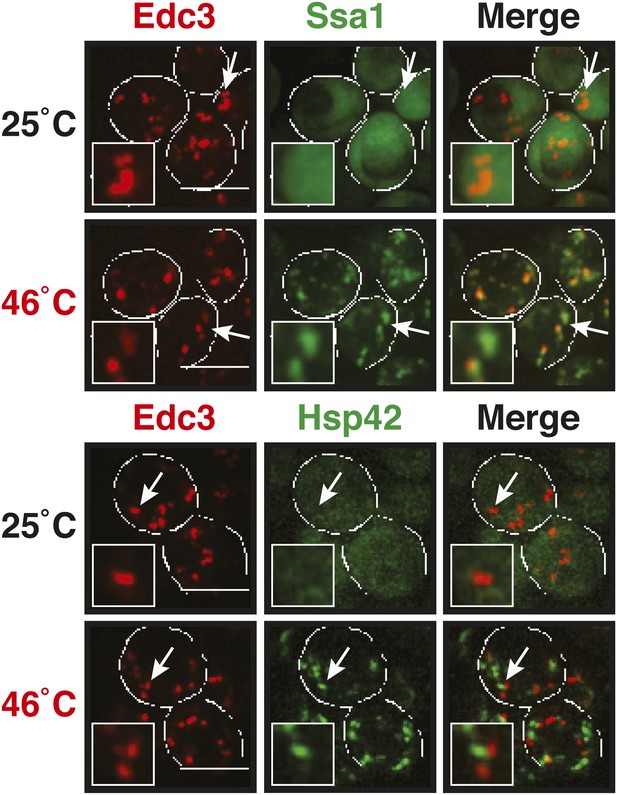
P bodies only show limited co-localization with chaperones under robust heat shock conditions.
GFP-tagged Hsp42 or Ssa1 was expressed from the endogenous locus and mCherry-tagged Edc3 from a plasmid. Cells were observed before and after heat shock at 46°C. White lines indicate the cell boundaries. Scale bars: 5 µm.
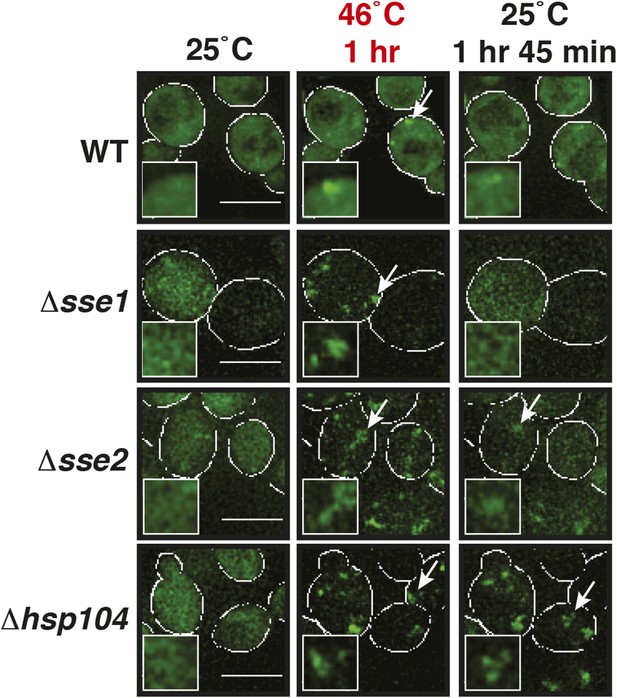
Lsm4-positive assemblies show different morphologies and behavior in the absence of disaggregases.
Fluorescence time-lapse microscopy of yeast cells expressing GFP-tagged Lsm4 from the endogenous locus. Wild-type cells are compared to strains lacking functional genes for disaggregases (Δhsp104, Δsse1, or Δsse2) after 1 hr heat stress at 46˚C and 1 hr 45 min after recovery at 25˚C. White lines indicate the cell boundaries. Scale bars: 5 µm.
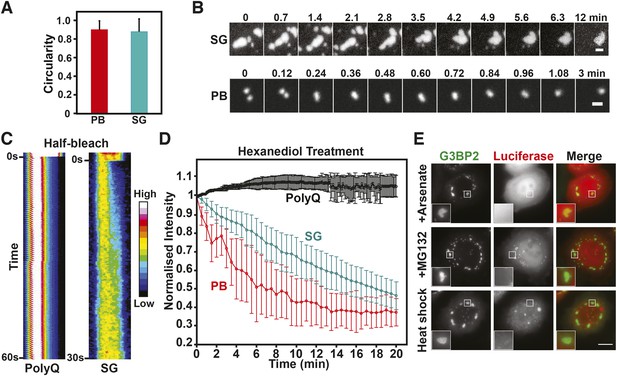
Mammalian RNP granules have liquid-like properties and are distinct from yeast stress granules.
(A) Mammalian P bodies and stress granules are spherical. Quantification of the circularity of stress granules (SG) and P bodies (PB) in HeLa cells, which expressed GFP-tagged DCP1a (PB) or G3BP2 (SG) from BACs (Poser et al., 2008). Cells were stressed with 1 mM sodium arsenate for 1 hr. Because of surface tension, liquid-like structures are expected to display a near circular shape (Hyman et al., 2014). A perfect circle has a circularity of 1. See ‘Materials and methods’ for details about how the circularity was determined. The mean and the SD are shown (n=118 PB, n=165 SG). (B) P bodies and stress granules show fusion behavior. Time-lapse microscopy of P bodies (DCP1a-GFP) and stress granules (G3BP2-GFP) in arsenate-stressed HeLa cells. Stress granules undergo frequent fusions at the beginning of a stress stimulus and form large spherical structures in cells after extended exposure to stress. Scale bars: 1 µm. Also see corresponding Videos 14, 15. (C) The stress granule protein G3BP2 shows fast internal rearrangement within stress granules, consistent with a liquid-like state. Kymograph of a stress granule (G3BP2-GFP) induced through arsenate stress and an amyloid (Q103-GFP) aggregate in HeLa cells after a half-bleach event (Brangwynne et al., 2009). PolyQ aggregates are used as a control for a solid-like structure. Note that only stress granules but not polyQ assemblies show a redistribution of fluorescence from the bleached to the unbleached area (from left to right). (D) P bodies and stress granules but not solid-like polyQ aggregates are sensitive to hexanediol. Quantification of the effect of 3.5% 1,6-hexanediol treatment on stress granules and P bodies induced through arsenate stress, and Q103 aggregates. The normalized mean fluorescence intensities of the structures are plotted over the duration of the treatment. Error bars are SD (n=5 PolyQ, n=31 SG and n=22 PB). (E) Mammalian stress granules do not co-aggregate with misfolded proteins. HeLa cells expressing BAC-encoded G3BP2-GFP (SG) were transfected with a plasmid coding for mutant luciferase-mCherry. Formation of stress granules was induced by arsenate stress (2 hr), proteasome inhibition (10 µM MG132, 3 hr) or heat stress (43°C, 2 hr). Scale bar: 10 µm.
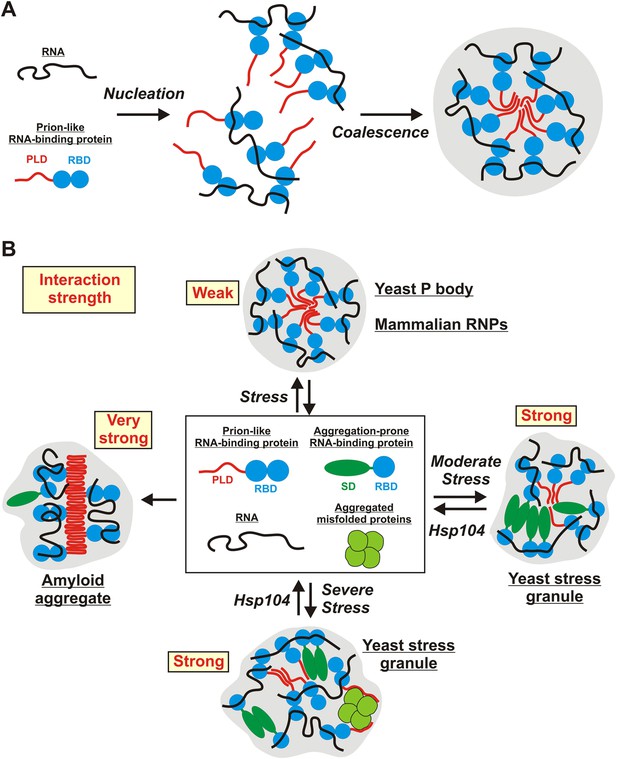
Schematic illustrating the potential molecular mechanisms underlying the formation of stress-inducible RNP granules.
(A) RNA-protein (RNP) granule formation as a two-step assembly process. In the first step, RNAs and RNA-binding proteins associate to form large RNP complexes (nucleation step). In the second step, these RNP complexes coalesce into larger compartments through additional RNA-mediated interactions, but primarily through PLD-mediated weak binding events (coalescence step). PLDs are indicated in red and RBDs indicated in blue. (B) Stress-inducible RNP granules have different compositions, which affect their dynamic and material properties. The presumed average interaction strength is indicated in red. Weak interactions increase the vulnerability to hexanediol. An aggregation-prone stress sensor domain (SD) is indicated in dark green.
Videos
Hexanediol treatment specifically disrupts non-amyloid Rnq1PD assemblies and not amyloids.
Fluorescence time-lapse microscopy of [PIN+] and [pin−] cells expressing Rnq1PD-sfGFP. All cells were treated with 10 µg/ml digitonin and, where indicated, with 10% 1,6-hexanediol. In the control condition only media was added. Related to Figure 1.
Reformation of non-amyloid Rnq1PD assemblies after hexanediol removal.
Cells expressing Rnq1PD-sfGFP were treated with 10% 1,6-hexanediol and 10 µg/ml digitonin for 1 hr to dissolve non-amyloid Rnq1PD assemblies. Hexanediol was washed out and replaced with normal growth media and the cells were observed by fluorescence time-lapse microscopy. Related to Figure 1.
P-body formation requires RNA.
Glucose starved cells containing Lsm4-GFP in the PB state or amyloid-like state (‘Prion’) were treated with 100 µg/ml cycloheximide and observed by fluorescence time-lapse microscopy. Related to Figure 2.
Hexanediol treatment disrupts P bodies in yeast cells.
Cells expressing Lsm4-GFP from the endogenous promoter were stressed in medium without glucose for 30 min. 5% hexanediol or medium (Control) was added and the cells were analyzed by time-lapse fluorescence microscopy. Related to Figure 3.
Reformation of P bodies after hexanediol removal.
Glucose starved cells expressing Lsm4-GFP were treated with 5% hexanediol for 1 hr. Hexanediol was washed out with fresh medium and the cells were observed by time-lapse fluorescence microscopy. Related to Figure 3.
Hexanediol does not disrupt stress granules.
Cells expressing Nrp1-GFP from the endogenous promoter were stressed for 15 min in glucose-deficient medium before the addition of 10% hexanediol. The cells were treated with 10 µg/ml digitonin to make them more permeable to hexanediol. The control received only medium +10 µg/ml digitonin. Note that P bodies dissolve within 2 min under the same conditions (see Figure 3—figure supplement 1). Related to Figure 4.
Stress granule and protein aggregate formation and dissolution in stressed cells.
Yeast cells expressing Nrp1-GFP from the endogenous locus were transformed with plasmids for the expression of mCherry-tagged mutant luciferase. Cells were exposed to a robust heat shock at 46°C and then transferred back to 25°C. Note that stress granules dissolve faster than protein aggregates. Related to Figure 5.
Stress granule and protein aggregate formation and dissolution in stressed cells.
Yeast cells expressing Nrp1-GFP from the endogenous locus were transformed with plasmids for the expression of mCherry-tagged Ubc9ts. Cells were exposed to a robust heat shock at 46°C and then transferred back to 25°C. Note that stress granules dissolve faster than protein aggregates. Related to Figure 5.
Stress granule and protein aggregate formation and dissolution in stressed cells.
Yeast cells expressing Pbp1-GFP from the endogenous locus were transformed with a plasmid for the expression of mCherry-tagged Ubc9ts. Cells were exposed to a robust heat shock at 46°C and then transferred back to 25°C. Note that stress granules dissolve faster than protein aggregates. Related to Figure 5.
The stress granule protein Nrp1 is not co-deposited with misfolding-prone proteins during glucose deprivation.
Fluorescence microscopy of yeast cells expressing Nrp1-GFP from the endogenous locus and mCherry-tagged mutants luciferase from a plasmid. Related to Figure 5.
Dissolution of stress granules is dependent on disaggregases.
Fluorescence time-lapse microscopy of yeast cells expressing GFP-tagged Nrp1 from the endogenous locus. Wild-type cells are compared to strains with genetic deficiencies (Δhsp104, Δsse1, or Δsse2). Cells were exposed to a robust heat shock at 46°C and then transferred back to 25°C. Related to Figure 5.
Edc3-positive assemblies show different morphologies and behavior in the absence of disaggregases.
Fluorescence time-lapse microscopy of yeast cells expressing GFP-tagged Edc3 from the endogenous locus. Wild-type cells are compared to strains lacking functional genes for certain disaggregases (Δhsp104, Δsse1 or Δsse2). Cells were exposed to a robust heat shock at 46°C and then transferred back to 25°C. Related to Figure 7.
Edc3 co-aggregates with luciferase in a Hsp104-dependent manner.
Endogenous hsp104 was deleted and substituted with plasmid-expressed Hsp104 under control of a GPD (high), ADH1 (medium), or SUP35 (low) promoter. Cells expressing mCherry-tagged luciferase from a plasmid and GFP-tagged Edc3 from the endogenous locus were exposed to a robust heat shock at 46°C and then transferred back to 25°C. Related to Figure 7.
Stress granules show extensive fusion behavior and form more spherical structures with time.
HeLa cells expressing BAC-encoded G3BP2 were observed by time-lapse microscopy. The cells were stressed through arsenate. Related to Figure 8.
Two P bodies fuse and rapidly relax into a spherical shape.
Time-lapse microscopy of P bodies (labelled by BAC-encoded DCP1a-GFP) in arsenate-stressed HeLa cells. Related to Figure 8.
Additional files
-
Supplementary file 1
List of yeast strains used in this study.
- https://doi.org/10.7554/eLife.06807.057
-
Supplementary file 2
Matlab routine for measuring the circularity of granules.
- https://doi.org/10.7554/eLife.06807.058





