A functional link between the co-translational protein translocation pathway and the UPR
Figures
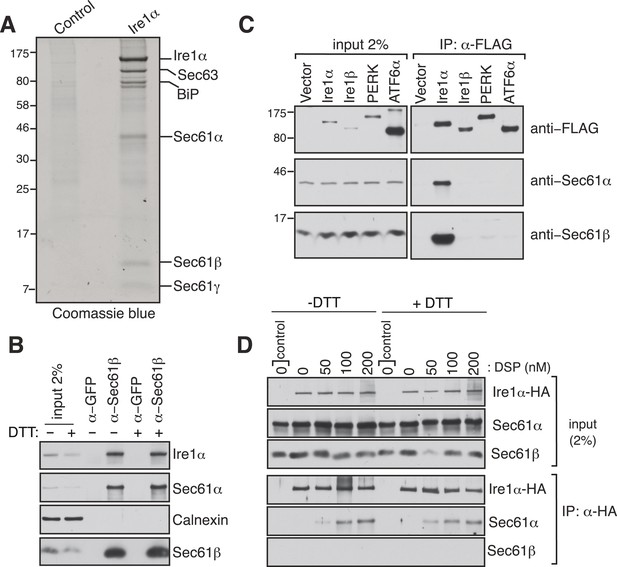
Identification of a complex between Ire1α and the Sec61 translocon.
(A) The detergent extracts of either microsomes derived from HEK 293 cells (control) or cells expressing hemagglutinin (HA)-tagged Ire1α were bound to anti-HA resin and eluted with a low pH glycine buffer. The eluted proteins were analyzed by SDS-PAGE and stained with coommassie blue. (B) The cell lysates from non-transfected HEK 293 cells treated with or without DTT were immunoprecipitated (IP) with anti-GFP antibodies as a control or anti-Sec61β antibodies. The bound material was eluted with sample buffer and analyzed along with starting lysates (input, 2% loading) by immunoblotting (IB) using antibodies against the indicated antigens. Calnexin, an abundant endoplasmic reticulum (ER) trans membrane protein was probed as a control. (C) Cell extracts from HEK 293 cells transfected with the indicated FLAG tagged constructs were subject to IP with FLAG antibody. The resulting samples were analyzed by IB with indicated antibodies. (D) HEK 293 cells stably expressing HA-tagged Ire1α were either treated with 10 mM DTT or left untreated for 2 hr. Cells were then semipermeabilized with 0.015% digitonin and treated with the indicated concentration of DSP crosslinker for 30 min at room temperature. Samples were denatured and IP with anti-HA antibodies. The resulting IP was analyzed by IB. Control denotes non-transfected HEK293 cells.

Peptides of Sec61α, Sec61β, and Sec61γ identified by mass spectrometry sequences of Sec61α, Sec61β, and Sec61γ annotated to indicate the peptides (yellow) identified by mass spectrometry.
https://doi.org/10.7554/eLife.07426.004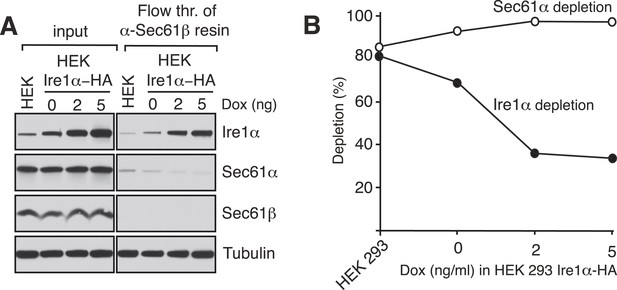
Ire1α is codepleted with the Sec61α translocon.
(A) The co-depletion of Ire1α with the Sec61 translocon was compared between endogenous Ire1α and recombinant Ire1α-HA. The cell lysates either from non-transfected HEK 293 cells or HEK 293 cells stably expressing various amount of Ire1-HA were passed through an anti-Sec61β resin to deplete the Sec61 translocon. The flow through fractions was analyzed along with starting lysates (input) by IB. (B) The co-depletion of Ire1α with the Sec61 translocon was quantified by ImageJ64 from the immunoblot shown in A. Note that nearly all endogenous Ire1α in HEK 293 cells was codepleted with the Sec61 translocon, whereas overexpression of Ire1α leads to freeunbound Ire1α, presumably due to a saturation of the Sec61 translocon binding sites.
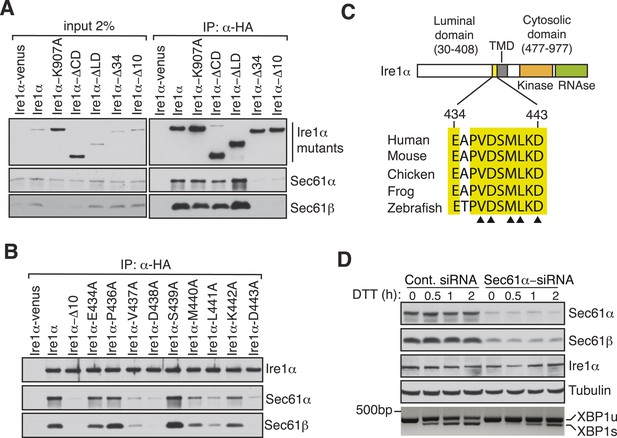
Key residues in Ire1α important for the interaction with the Sec61 translocon.
(A) The cell lysates of the indicated versions of HA-tagged Ire1α were IP with anti-HA antibodies, eluted with sample buffer and analyzed by IB. Ire1α–venus served as a control. The mutation K907 to A907 impairs the RNase activity of Ire1α (Tirasophon et al., 2000). Deletion of the Ire1α cytosolic domain from amino acid 477 to 977 or luminal domain from amino acid 30 to 408 is labeled as ΔCD or ΔLD (Volmer et al., 2013), respectively. Ire1α Δ34 carry a deletion from amino acid 409 to 443 and Ire1α Δ10 lacks amino acids 434 to 443. (B) The indicated Ire1α mutants were analyzed as described in panel A. (C) Comparison of the sequences of the 10 amino acid region of Ire1α in vertebrates. Triangle depicts amino acid residues of Ire1α in which alanine scanning mutations disrupt binding to the Sec61 translocon. (D) HeLa cells were transfected with control siRNA or siRNA targeting Sec61α. After 48 hr of transfection, cells were transfected again with siRNA which was followed by transfection with FLAG-tagged XBP1u. 96 hr after the first transfection, cells were treated with 10 mM DTT for the indicated time periods. Total proteins and RNA were isolated from Trizol harvested cells and analyzed by IB against the indicated antigens and by an RT-PCR reaction to monitor splicing of XBP1u mRNA (Calfon et al., 2002), respectively.
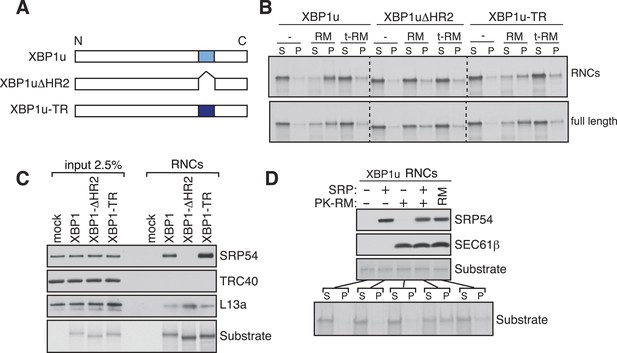
XBP1u utilizes the signal recognition particle (SRP) pathway for targeting its mRNA to the ER membrane.
(A) Diagram of constructs derived from XBP1u. Blue box denotes the previously described hydrophobic region 2 (HR2) of XBP1u (Yanagitani et al., 2009). Dark blue indicates the transmembrane domain (TMD) from the transferrin receptor in lieu of HR2. (B) The indicated versions of XBP1u transcripts lacking a termination codon were translated in rabbit reticulocyte lysate in the presence of rough microsomes (RM) or trypsin digested RM (tRM). The reactions were separated by centrifugation to analyze pellet (P) and soluble fractions (S) by SDS-PAGE and autoradiography. (C) Affinity purified ribosome associated nascent chains (RNCs) of the indicated versions of FLAG-tagged XBP1u were analyzed by IB for the indicated antigens. L13a is a ribosomal protein. TRC40 is a control protein. Autoradiography of the blot revealed equal recovery of translated substrates. (D) XBP1u transcripts lacking a termination codon were translated in the wheat germ translation system including purified SRP, puromycin/potassium acetate treated RM (PK-RM) or both. RM alone was included as a control. An aliquot of the total translation reaction was analyzed by IB for SRP54 and Sec61β, which indicate the presence of SRP and PK-RM, respectively. An autoradiograph of the blot revealed equal translation of substrate XBP1u in all reactions. The reactions were separated and analyzed as in panel B.
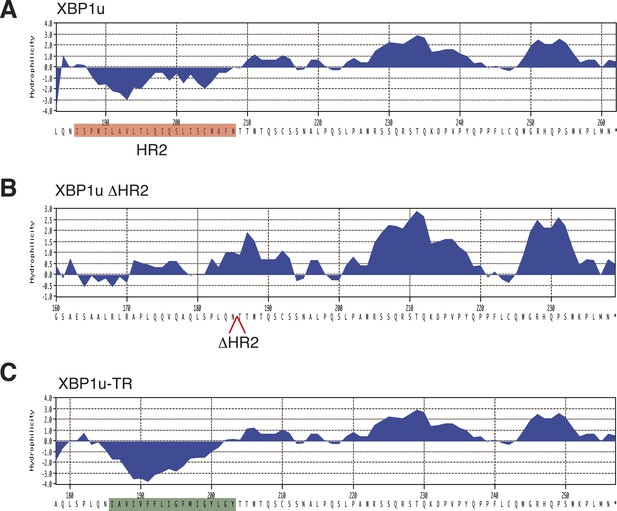
Sequence and hydrophobicity of XBP1u constructs.
The last 80 residues of the indicated XBP1u constructs used in this study were analyzed for hydrophobicity using the Kyte-Doolittle scale. (A) The red color denotes HR2 in XBP1u. (B) The HR2 deleted position is indicated as an open arrow. (C) The green color denotes the replacement of HR2 with the TMD from the transferrin receptor.
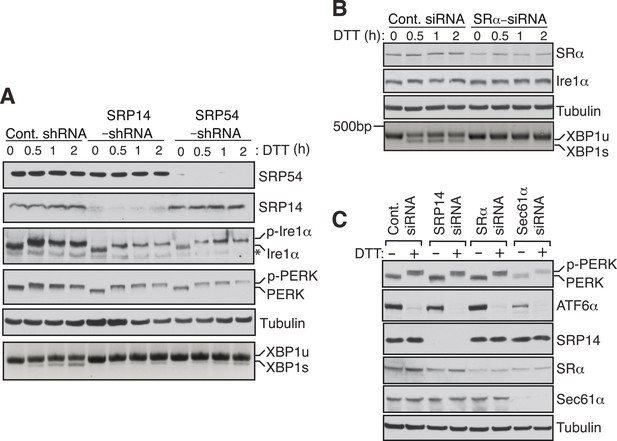
SRP mediated targeting to the ER ensures efficient spicing of XBP1u mRNA.
(A) HEK 293 cells were transfected with shRNAs against luciferase (control), SRP14, or SRP54. 5 days after transfection, the cells were replated for transfection with XBP1u and treated with 10 mM DTT for the indicated time periods. The Trizol harvested cells were analyzed as in Figure 2D. A phos-tag gel was used for the Ire1α immunoblot. p-Ire1α and p-PERK indicate the phosphorylated forms of Ire1α and PERK, respectively. * denotes a background band. (B) HeLa cells were transfected with control siRNA or siRNA targeting the α subunit of the SRP receptor (SRα) and analyzed as described in Figure 2D. (C) HeLa cells were transfected with the indicated siRNA oligos and treated with 10 mM DTT for 2 hr after 96 hr post-transfection. Cells were harvested in SDS sample buffer and analyzed for IB with indicated antibodies. Upon DTT treatment, the ATF6α band disappears due to the cleavage of its N-terminal cytosolic domain. Our ATF6α antibodies were not suitable for detecting the cleaved N-terminal cytosolic domain (not shown). Note that depletion of Sec61α caused significant reduction of the transmembrane proteins PERK and ATF6α.
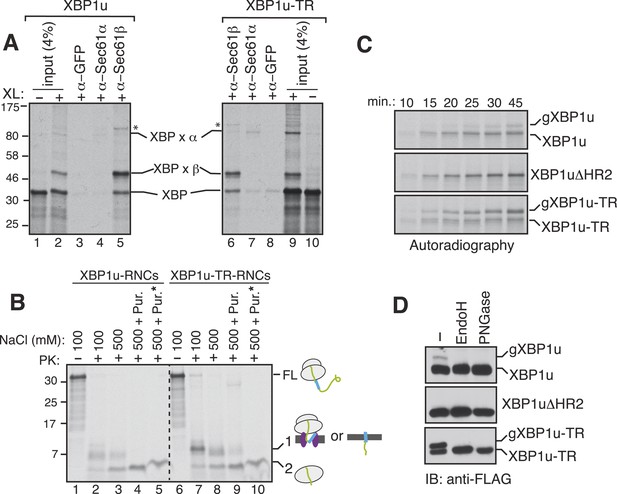
XBP1u nascent chains interact with the Sec61 translocon, but inefficiently insert into the ER membrane.
(A) The membrane-targeted RNCs of XBP1u or XBP1u-TR were isolated by centrifugation and treated with BMH crosslinker. An aliquot was directly analyzed (input, 4% loading), while the remainder was IP with the indicated antibodies. Anti-GFP antibodies were used as a control. The XBP1u crosslinked adducts are indicated by ‘XBP x’. * indicates an unidentified crosslinked product. (B) The ER membrane targeted RNCs of XBP1u variants were subjected to a proteinase K (PK) accessibility assay in the presence of the indicated salt concentrations and salt plus puromycin (pur.). * indicates the inclusion of a detergent in the reaction. FL indicates full-length versions of XBP1u. Band 1 indicates protease-protected fragments of either ribosome translocon protected fragments or protected fragments after insertion into the membrane (lane 9). Band 2 indicates fragments protected by ribosomes. (C) The indicated versions of XBP1u transcripts containing a glycan acceptor site at the C-terminus were translated in vitro in the presence of RM. The reactions were stopped at the indicated time points by directly mixing with the sample buffer and analyzed by autoradiography. gXBP1u denotes the glycosylated form. (D) Cell lysates from HEK 293 cells expressing the indicated FLAG tagged XBP1u versions containing a glycan acceptor site were treated with endoglycosidase H (EndoH) or peptide-N-glycosidase F (PNGase) and analyzed by IB with FLAG.
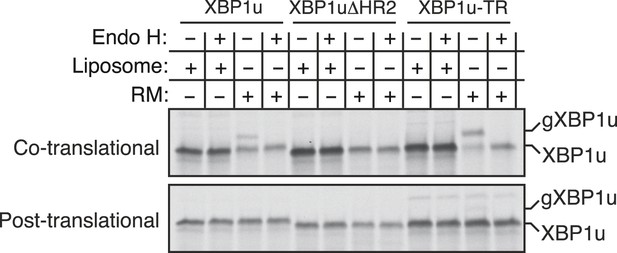
Insertion assays with XBP1u and its variants.
For the co-translational protein insertion, versions of XBP1 transcripts containing a glycan acceptor site were translated in vitro in the presence of liposomes or RM. The reactions were stopped after 30 min by directly mixing with the endoglycosidase H (EndoH) denaturation buffer and digested with Endo H before analyzing by autoradiography. gXBP1u denotes the glycosylated form of XBP1u. For the post-translational protein insertion, the indicated XBP1u transcripts were initially translated in the absence of RM and the nascent chains were released by the puromycin treatment. The post ribosomal supernatant was used for the insertion assay.
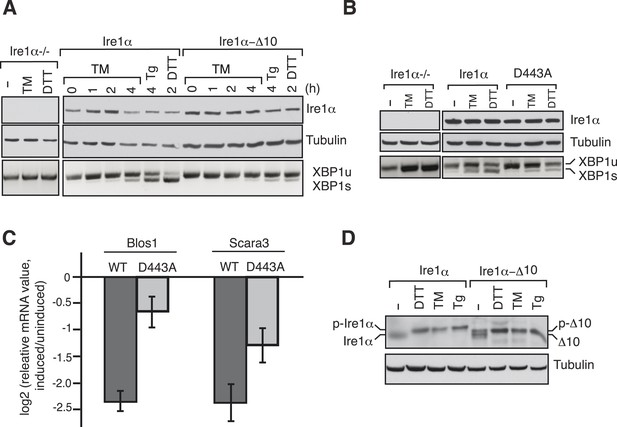
The Ire1α interaction with the Sec61 translocon ensures efficient cleavage of ER-targeted mRNAs.
(A) HEK 293 Ire1α−/− cells generated by CRISPR/Cas9 were stably complemented with Ire1α-HA or its mutant (Δ10). The expression of these constructs was controlled by doxycycline, but the cells were not induced with doxycyline in order to achieve low expression levels of Ire1α. Cells were harvested in Trizol after either treatment with tunicamycin (TM: 5 μg/ml), thapsigargin (Tg: 2.5 μg/ml) or DTT (10 mM) for the indicated time periods and analyzed by XBP1u mRNA splicing assay and IB with the indicated antibodies. (B) Mouse embryonic fibroblast (MEF) Ire1α−/− cells complemented with Ire1α-HA or its mutant (D443A) were harvested after either treatment with TM (5 μg/ml) for 5 hr or DTT (10 mM) for 2 hr and analyzed by XBP1u mRNA splicing assay and IB as described in Figure 2D. (C) The MEF Ire1α−/− cells complemented with indicated Ire1α variants were treated with TM (5 μg/ml) for 6 hr and analyzed by qPCR to measure Blos1 and Scara3 mRNA abundance. We normalized all mRNA abundance measurements to the housekeeping control Rpl19 mRNA. (D) HEK 293 Ire1α−/− cells stably expressing Ire1α-HA or its mutant (Δ10) were treated with DTT for 2 hr, TM for 5 hr, Tg for 5 hr and analyzed for phosphorylated Ire1α.
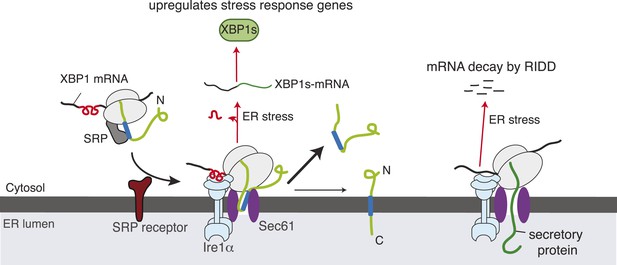
Model for Ire1α-mediated cleavage of ER-localized mRNAs.
Ire1α forms a complex with the Sec61 translocon, to which XBP1u mRNA is recruited by its ribosome nascent chains (RNCs) through the SRP pathway. Despite interacting with the Sec61 translocon, the XBP1u nascent chain is inefficiently inserted into the ER membrane due to its weak hydrophobic region. Upon ER stress, Ire1α is activated through self-oligomerization and cleaves XBP1u mRNA to yield an active transcription factor, XBP1s, as well as to cleave ER-localized mRNAs through regulated Ire1-dependent decay (RIDD).
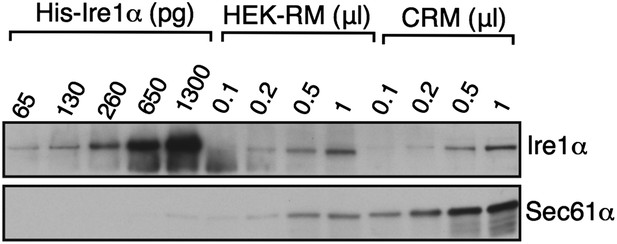
Ire1α concentration in HEK 293 microsomes.
Ire1α concentration in either HEK 293 microsomes or canine microsomes (CRM) was quantified relative to purified His-tagged Ire1α. We estimate 1μl of HEK microsomes (OD280 = 50) contain ∼150 pg or 1.4 nM of Ire1α. Tyedmers et al. (2000, PNAS) estimated that 1ul of CRM (OD280 = 50) contains ∼ 2.12 uM. Thus, 1 μl of HEK microsomes contain ∼ 0.4 uM since they contain 5X less concentration of Sec61. We therefore estimate that Ire1α concentration is ∼ 300X less than the Sec61 in HEK microsomes. Note that the five times less concentration of Sec61 in HEK relative to CRM is not likely biased by our Sec61α antibodies since both human and canine Sec61α peptide sequence are nearly identical.





