Centrosome age regulates kinetochore–microtubule stability and biases chromosome mis-segregation
Figures
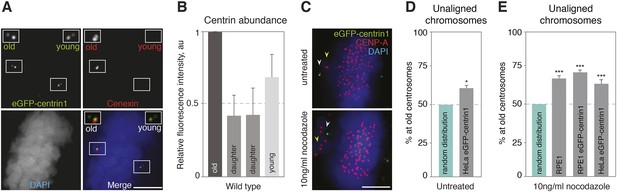
Centrosome age affects chromosome alignment.
(A) HeLa-eGFP-centrin1 (green) cell stained for cenexin (red, old centrosome marker) and DAPI (blue, DNA). One spindle pole contains the old centriole (brightest centrin1 signal and cenexin positive) and an accompanying daughter centriole (dim signal), which together form the old centrosome. The other spindle pole contains the young centriole (intermediate centrin1 signal), which is also accompanied by a daughter centriole and which together form the young centrosome. Scale bar in all panels = 5 μm. (B) Amounts of eGFP-centrin1 on the old, young and daughter centrioles in HeLa-eGFP-centrin1 cells determined from 3 independent experiments in 140 cells. (C) Untreated HeLa-eGFP-centrin1 cell (upper panel) and hTert RPE-eGFP-centrin1 cell treated with 10 ng/ml nocodazole (lower panel) stained for CENP-A (kinetochore marker) and DAPI. Yellow arrowheads indicate unaligned chromosomes; white arrowheads old centrosomes. (D and E) Proportion of unaligned chromosomes at old centrosomes in HeLa-eGFP-centrin1 cells (D), and in RPE1 cells stained for cenexin, RPE1-eGFP-centrin1 cells and HeLa-eGFP-centrin1 cells treated with 10 ng/ml nocodazole (E). For experiment and cell numbers, and p-values see Table 1. For results of individual experiments see Figure 1—source data 1. Error bars indicate s.e.m. * indicates p ≤ 0.05 in Binomial test compared to random distribution, *** indicates p ≤ 0.01 in Binomial test compared to random distribution.
-
Figure 1—source data 1
Values of individual experiments of graphs shown in Figure 1.
- https://doi.org/10.7554/eLife.07909.004
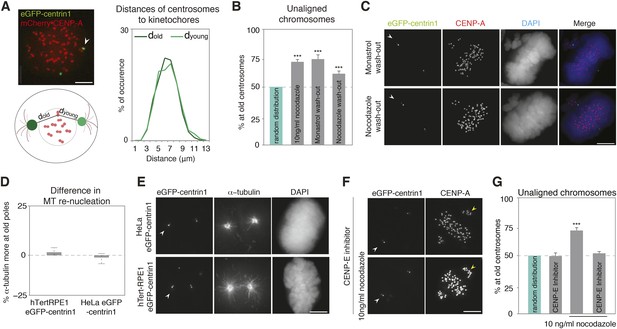
The asymmetric distribution of unaligned chromosomes depends on CENP-E.
(A) An illustrative example of a HeLa-eGFP-centrin1/mCherry-CENP-A cell where the distances from centrosomes to kinetochores were measured 30 s before nuclear envelope breakdown (left top), assay to calculate the distances between kinetochores and centrosomes (left bottom), and distribution (right) of the measured distances. Values are determined from 24 cells and 1434 kinetochores in 6 independent experiments. White arrowheads indicate old centrosomes in all panels. Scale bars in all panels = 5 μm. (B) Proportion of unaligned chromosomes at the old centrosomes in hTert-RPE1-eGFP-centrin1 cells after indicated treatments. Error bars indicate s.e.m. *** indicates p ≤ 0.01 in Binomial test. (C) hTert-RPE1-eGFP-centrin1 cells stained for CENP-A and DAPI after indicated treatments. (D) Differences in the intensity of the microtubule asters in a re-nucleation assay at old and young centrosomes as shown in E, calculated in 32–49 cells in 3 independent experiments. Columns indicate the median, errors bars the 99% CI. Precise methodology is shown in Figure 2—figure supplement 2. (E) HeLa-eGFP-centrin1 or hTert-RPE1-eGFP-centrin1 cells stained for α-tubulin after a microtubule re-nucleation assay. (F) hTert-RPE1-eGFP-centrin1 cells treated with 10 ng/ml nocodazole and/or CENP-E inhibitor, and stained for CENP-A. Yellow arrowheads indicate unaligned kinetochores in the proximity of the young centrosome (G) Proportion of unaligned chromosomes at old centrosomes in hTert-RPE1-eGFP-centrin1 cells treated with 10 ng/ml nocodazole and/or CENP-E inhibitor. Error bars indicate s.e.m. *** indicates p ≤ 0.01 in Binomial test. For results of all individual experiments see Figure 2—source data 1.
-
Figure 2—source data 1
Values of individual experiments of graphs shown in Figure 2.
- https://doi.org/10.7554/eLife.07909.007
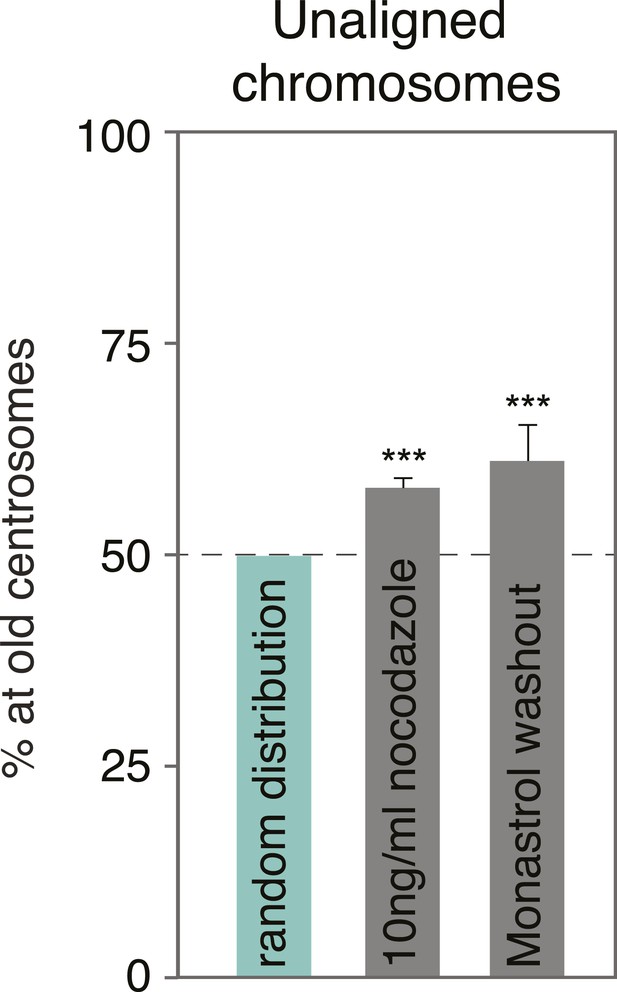
Monastrol wash-out does not change the alignment bias in HeLa cells.
Proportion of unaligned chromosomes at the old centrosome in HeLa-eGFP-centrin1 cells after monastrol washout. Errors bars indicate s.e.m. *** indicates p ≤ 0.01 in Binomial test.
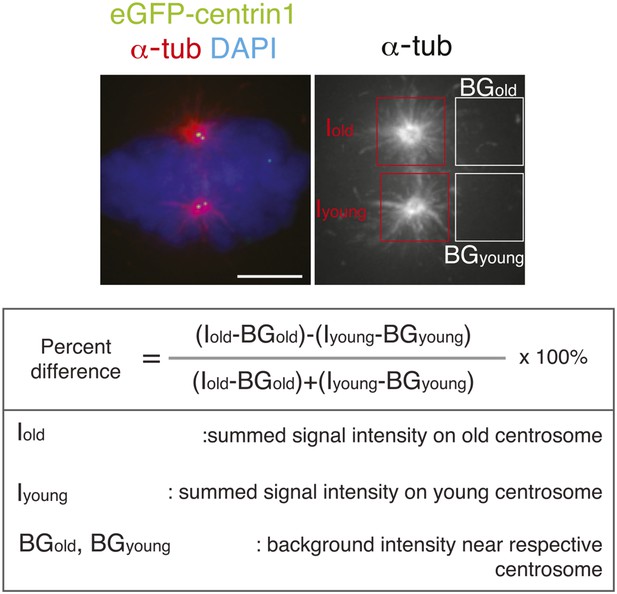
Methodology to compare microtubule re-nucleation at old and new pole.
To calculate in hTert-RPE1-eGFP-centrin1 cells the relative differences in microtubule nucleation, cells were stained after a release from an ice-cold treatment with a-tubulin antibodies (microtubules) and DAPI (chromosomes). The intensity of the centrosomal aster (minus the background) at old and young centrosomes was measured and the relative differences calculated with the indicated formula. Scale bar = 5 μm.
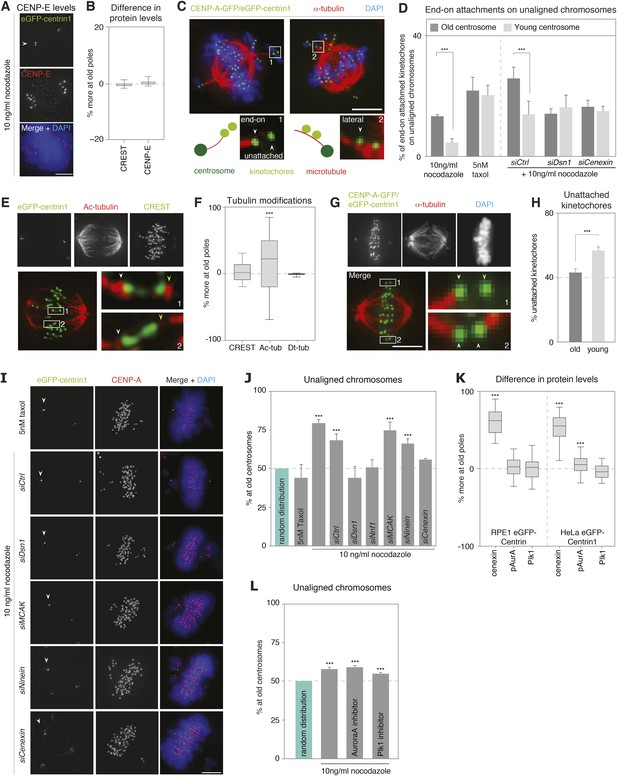
Kinetochore–microtubules bound to old centrosomes are more stable.
(A) hTert-RPE1-eGFP-centrin1 cells treated with 10 ng/ml nocodazole and stained for CENP-E and DAPI. White arrowheads indicate old centrosomes in all panels. Scale bars in all panels = 5 μm. (B) Differences in the abundance of CENP-E and CREST (kinetochore marker) at kinetochores bound to old and young centrosomes, calculated from 27 cells in 3 independent experiments. Columns indicate the median; error bars the 99% CI. (C) Immunofluorescence image of a HeLa-eGFP-centrin1/eGFP-CENP-A (green) cells treated with 10 ng/ml nocodazole, fixed with glutaraldehyde, and stained for α-tubulin (red) and DAPI (blue). Single kinetochores in every unaligned sister-kinetochore pair were classified as end-on attached, laterally attached or unattached. Inset 1 on the left shows an illustrative example of a kinetochore pair with one unattached and one end-on attached kinetochore; inset 2 on the right shows an illustrative example with 2 laterally attached kinetochores. (D) Quantification of individual end-on attached kinetochores at old and young centrosomes in HeLa-eGFP-centrin1/eGFP-CENP-A cells treated with 10 ng/ml nocodazole, 5 nM taxol and the indicated siRNAs and 10 ng/ml nocodazole. Percentages are based on 3 independent experiments with 29–50 cells. Error bars indicate s.e.m; *** indicates p ≤ 0.01 in paired t-test. (E) hTertRPE1-eGFP-centrin1 cells stained with anti-acetylated tubulin (red) and CREST (green) antibodies. Shown are total projections (upper panels) or maximum-intensity projections of 5–10 planes around the focal plane of interest (lower panels). White arrowheads indicate kinetochore–microtubules with stronger acetylation, yellow with weaker acetylation. Note that the white arrows are on the side of the old centrosome. (F) Differences in the abundance of acetylated tubulin on k-fibres of sister-kinetochores, and detyrosinated tubulin on the two spindle halves in hTertRPE1-eGFP-centrin1 cells, based on 3 independent experiments and 32–33 cells. Methodology is explained in Figure 3—figure supplement 2. Columns indicate the meadian, error bars the 99% CI. (G) HeLa-eGFP-centrin1/eGFP-CENP-A cells treated with 0.5 mM Ca2+ for 10 min stained for α-tubulin (red) and DAPI (blue). Shown are total projections (upper panels) or maximum-intensity projections of 5–10 planes around the focal plane of interest (lower panels). White arrowheads in zoom-ins indicate end-on attached kinetochores and yellow arrow the unattached kinetochore. (H) Percentage of unattached kinetochores oriented towards old or young poles based on 3 independent experiments and 33 cells. (I) hTert-RPE1-eGFP-centrin1 cells stained with CENP-A antibodies (red) and DAPI (blue) after treatment with 5 nM taxol or the indicated siRNAs and 10 ng/ml nocodazole. White arrowheads indicate old centrosome. (J) Proportion of unaligned chromosomes at old centrosome in hTert-RPE1-eGFP-centrin1 cells treated with 5 nM taxol or with 10 ng/ml nocodazole after the indicated siRNA treatment. Error bars indicate s.e.m; *** indicates p ≤ 0.01 in Binomial test. (K) Differences in the abundance of cenexin, phospho-Aurora-A, and Plk1 at old and young centrosomes in HeLa and hTert-RPE1-eGFP-centrin1 cells, based on 3 independent experiments and 41–113 cells. Methodology is explained in Figure 3—figure supplement 2. Columns indicate the median, error bars the 99% CI. (L) Proportion of unaligned chromosomes at old centrosome in HeLa-eGFP-centrin1 cells treated with Aurora-A or Plk1 inhibitors. Error bars indicate s.e.m; *** indicates p ≤ 0.01 in Binomial test. For results of all individual experiments see Figure 3—source data 1.
-
Figure 3—source data 1
Values of individual experiments of graphs shown in Figure 3.
- https://doi.org/10.7554/eLife.07909.011
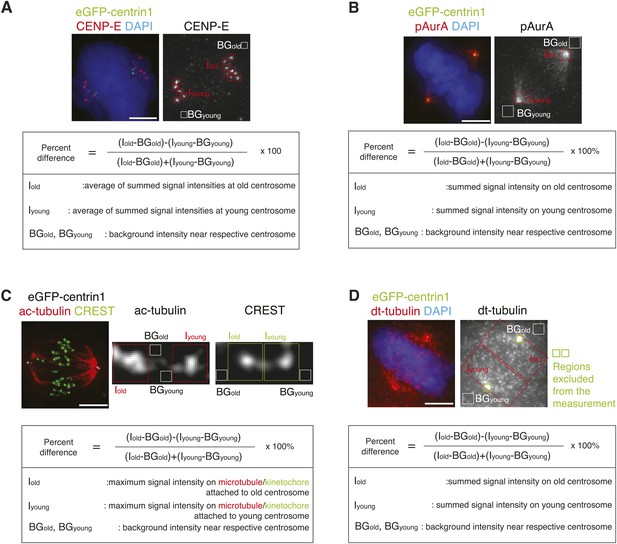
Methodology to compare kinetochore- and centrosome-associated protein intensities at old and young spindle poles.
(A) To calculate in hTert-RPE1-eGFP-centrin1 cells the relative differences in the abundance of CENP-E on unaligned kinetochores, cells were stained with CENP-E antibodies and DAPI (chromosomes). The intensity of CENP-E on unaligned kinetochores (minus the background) at old and young centrosomes was measured and the relative differences calculated with the indicated formula. Scale bars in all panels = 5 μm. (B) Methodology to calculate the relative differences in phospho-Aurora-A, Plk1, or cenexin in hTert-RPE1-eGFP-centrin1 cells as displayed in Figure 3K. In this example, cells were stained with phospho-Aurora-A antibodies, before determining its abundance on old and young centrosomes. The relative difference was calculated with the indicated formula. (C) Methodology to calculate the relative differences in acetylated tubulin (ac-tubulin) in hTert-RPE1-eGFP-centrin1 cells as displayed in Figure 3E,F. In this example cells were stained with acetylated tubulin antibodies, before determining its abundance on the kinetochore–microtubules associated with old and young centrosomes. The relative difference was calculated with the indicated formula. (D) Methodology to calculate the relative differences in detyrosinated tubulin (dt-tubulin) in hTert-RPE1-eGFP-centrin1 cells as displayed in Figure 3F. Cells were stained with detyrosinated tubulin antibodies, before determining its abundance on the half-spindles associated with old and young centrosomes. Same area around the old and the young centrosome, containing centrioles, was excluded from the measurement (green rectangles). The relative difference was calculated with the indicated formula.
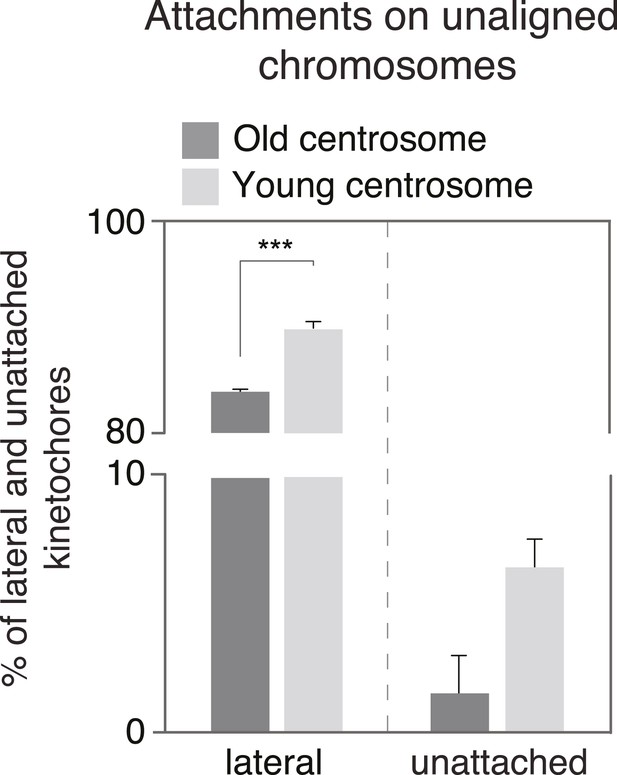
Quantification of the proportion of laterally and unattached kinetochores.
Proportion of laterally attached (left) or unattached kinetochores (right) at old and young centrosomes in HeLa-eGFP-centrin1/eGFP-CENP-A cells treated with 10 ng/ml nocodazole, as determined in immunofluorescence pictures shown in Figure 3C.
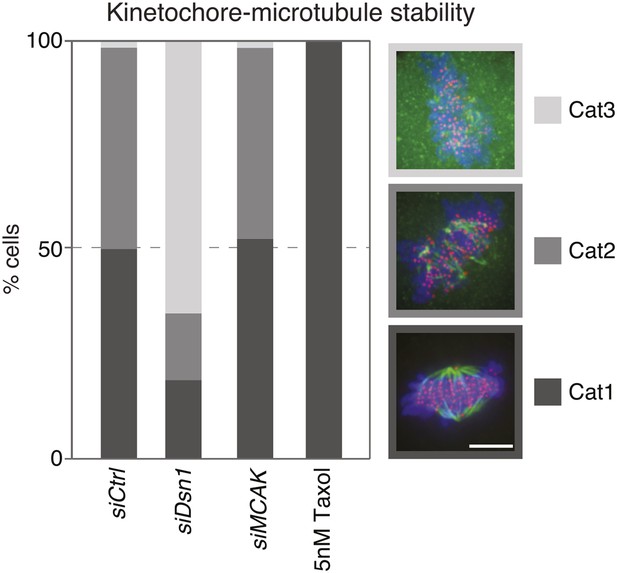
Quantification of kinetochore–microtubule stability in cold treated cells.
The bar graph indicates the percentage of HeLa-eGFP-centrin1/eGFP-CENP-A cells treated with the indicated siRNAs or taxol displaying intact kinetochore–microtubules(Category 1), destabilized kinetochore–microtubules (Category 2) or no kinetochore–microtubules (Category 3), after 20 min of cold treatment on ice. Values are based on measurements in 50–60 cells in 3 independent experiments. Scale bar = 5 µm.
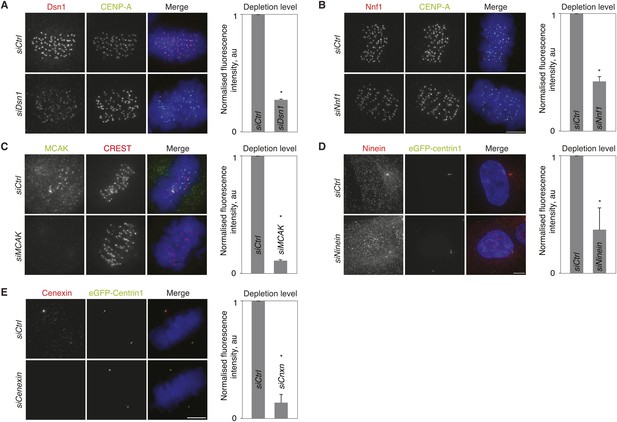
Validation of siRNA treatments.
(A–E) hTert-RPE1-eGFP-centrin1 cells treated with Dsn1 (A), Nnf1 (B), MCAK (C), Ninein (D) or Cenexin (E) siRNA and stained with indicated antibodies. Note that ninein levels were quantified in interphase, as ninein is barely visible at centrosomes in mitotic cells. Scale bar in all panels = 5 μm. Errors bars indicate s.e.m. * indicates p ≤ 0.05 in paired t-test.
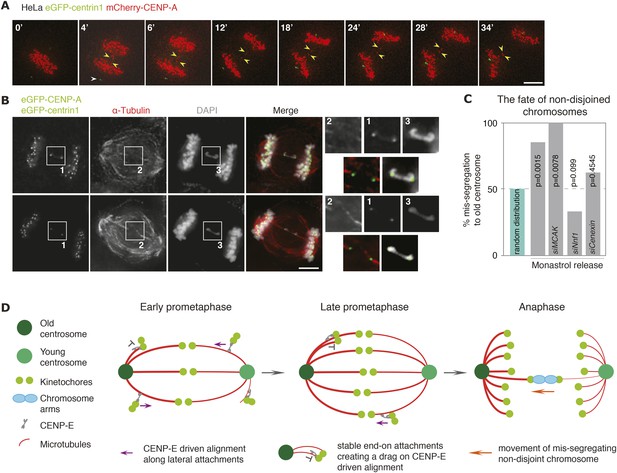
Non-disjoined chromosomes co-segregate with old centrosomes.
(A) Time lapse images of a HeLa-eGFP-centrin1/mCherry-CENP-A cell with a non-disjoined sister-kinetochore pair in anaphase. White arrowhead indicates the old centrosome, yellow arrowheads the non-disjoined sister-kinetochore pair. Scale bar = 10 μm. (B) Illustrative example of a HeLa-eGFP-centrin1/eGFP-CENP-A cell in anaphase stained for α-tubulin with a non-disjoined chromosome. Insets highlight the non-disjoined chromosomes. Scale bar = 5 μm. (C) Proportion of non-disjoined chromosomes that co-segregate with the old centrosomes in HeLa-eGFP-centrin1/mCherry-CENP-A cells treated with the indicated siRNAs. For statistics and number of experiments, see Table 2. (D) Proposed model of how old and new centrosomes differentially affect chromosome alignment and chromosome segregation via kinetochore–microtubule stability.
Videos
HeLa cell expressing mCherry-CENP-A (kinetochore marker in red) and eGFP-centrin1 (centrosome age marker in green) after a monastrol washout.
Note that the mis-segregating chromosome moves towards the brighter, old centrosome.
HeLa cell expressing mCherry-CENP-A (kinetochore marker in red) and eGFP-centrin1 (centrosome age marker in green) depleted of Nnf1, after a monastrol washout.
Note that the mis-segregating chromosome moves towards the dimmer, young centrosome.
HeLa cell expressing mCherry-CENP-A (kinetochore marker in red) and eGFP-centrin1 (centrosome age marker in green) depleted of Cenexin, after a monastrol washout.
Note that the mis-segregating chromosome moves towards the dimmer, young centrosome.
Tables
Percentage of unaligned chromosomes at old centrosomes
| Condition | N0 of repeats | N0 of cells | N0 of chromosomes | % Chromosomes at old centrosomes | 2-tailed Binomial test p |
|---|---|---|---|---|---|
| HeLa-eGFP-centrin1 DMSO | 3 | 33 | 93 | 61.23 | 0.037 |
| hTert-RPE1 10 ng/ml nocodazole | 3 | 161 | 227 | 67.80 | 8.08e-8 |
| hTert-RPE1-eGFP-centrin1 10 ng/ml nocodazole | 7 | 127 | 295 | 71.81 | 1.0e-12 |
| HeLa-eGFP-centrin1 10 ng/ml nocodazole | 3 | 57 | 532 | 63.91 | 1.42e-10 |
| hTert-RPE1 Eg5 inhibition recovery | 3 | 53 | 156 | 74.36 | 8.9e-10 |
| hTert-RPE1 nocodazole recovery | 3 | 59 | 164 | 61.58 | 0.00373 |
| HeLa-eGFP-CENP-A/eGFP-centrin1 10 ng/ml nocodazole | 3 | 142 | 946 | 58.03 | 8.68e-07 |
| HeLa-eGFP-CENP-A/eGFP-centrin1 Eg5 inhibition recovery | 5 | 68 | 306 | 61.11 | 0.000121 |
| hTert-RPE1-eGFP-centrin1 siCtrl 10 ng/ml nocodazole | 3 | 92 | 206 | 68.45 | 1.26e-07 |
| hTert-RPE1-eGFP-centrin1 siNinein 10 ng/ml nocodazole | 4 | 77 | 169 | 66.86 | 0.0000138 |
| hTert-RPE1-eGFP-centrin1 CENP-E inhibitor | 3 | 65 | 393 | 49.87 | n.s* |
| hTert-RPE1-eGFP-centrin1 CENP-E inhibitor 10 ng/ml nocodazole | 3 | 59 | 459 | 52.29 | n.s* |
| hTert RPE-eGFP-centrin1 5 nM Taxol | 3 | 50 | 105 | 43.81 | n.s* |
| hTert RPE-eGFP-centrin1 siDsn1 10 ng/ml nocodazole | 3 | 42 | 162 | 43.82 | n.s* |
| hTert RPE-eGFP-centrin1 siNnf1 10 ng/ml nocodazole | 5 | 21 | 46 | 52.17 | n.s* |
-
*
Non-significant.
Percentage of mis-segregating chromosomes that co-segregate with the old centrosomes
| Condition | N0 of repeats | No of cells | No of chromosomes | No of chromosomes to the old centrosome | 2-tailed binomial test p |
|---|---|---|---|---|---|
| HeLa-eGFP-centrin1/ mCherry-CENP-A | 11 | 21 | 21 | 18 | 0.0015 |
| HeLa-eGFP-centrin1/ mCherry-CENP-A siMCAK | 3 | 8 | 8 | 8 | 0.0078 |
| HeLa-eGFP-centrin1/ mCherry-CENP-A siNnf1 | 6 | 30 | 30 | 10 | 0.0990 |
| HeLa-eGFP-centrin1/ mCherry-CENP-A siCenexin | 6 | 16 | 16 | 10 | 0.4545 |
Additional files
-
Source code 1
Source code to calculate the distance between kinetochores and centrosomes at nuclear envelope breakdown
- https://doi.org/10.7554/eLife.07909.021





