An extracellular biochemical screen reveals that FLRTs and Unc5s mediate neuronal subtype recognition in the retina
Figures
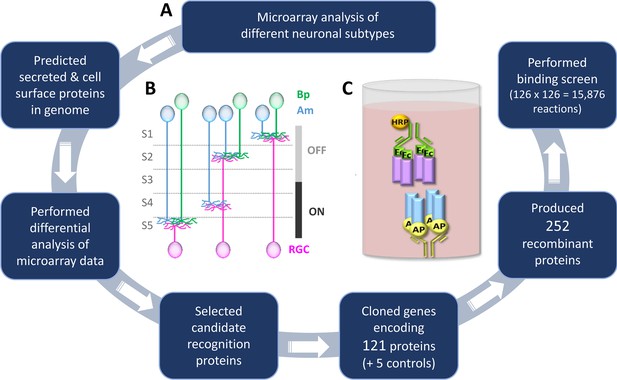
Methodology to identify recognition proteins for an extracellular receptor-ligand binding screen.
(A) Flow chart describing the process of conducting candidate-based binding screen. A flow chart depicting the process of predicting the cell surface and secreted proteins in the mouse genome prior to candidate selection is outlined in Figure 1—figure supplement 1. A table of the 65 candidate genes is included as Figure 1—source data 1 and a description of the 15 previously-unreported cDNAs that encode new isoforms is presented as Figure 1—source data 2. (B) Schematic representation of the IPL showing the five sublayers (S1-S5), three major classes of neurons: amacrines (Am, blue), bipolars (Bp, green), retinal ganglion cells (RGCs, magenta) and the function of the sublayers in visual processing (OFF and ON). Neurite stratifications provide an example of differential laminar organization. (C) Schematic representation of the ELISA-based binding assay. Receptor proteins (blue) tagged with alkaline phosphatase (AP; yellow) are tetramerized on the ELISA plate via an anti-AP antibody (yellow). Binding of tetramerized ligand (purple) tagged with the Fc region of IgG1 (Fc; green) to receptor is detected by inclusion of an anti-Fc antibody conjugated with horseradish peroxidase (HRP; orange).
-
Figure 1—source data 1
Table lists the 65 candidate genes selected for the binding screen, the 121 proteins encoded by different isoforms or cleavage products, EntrezGene identifiers and Accession numbers, primer sequences used for cDNA cloning of the extracellular domain, protein type (secreted, GPI-linked or transmembrane) and the protein concentrations for both the AP- and Fc-tagged proteins used in the binding screen.
- https://doi.org/10.7554/eLife.08149.004
-
Figure 1—source data 2
Previously-unreported cDNAs encoding new isoforms.
Table lists the gene symbols, the name assigned to each new isoform and a description of how the new isoform differs from previously reported cDNAs.
- https://doi.org/10.7554/eLife.08149.005
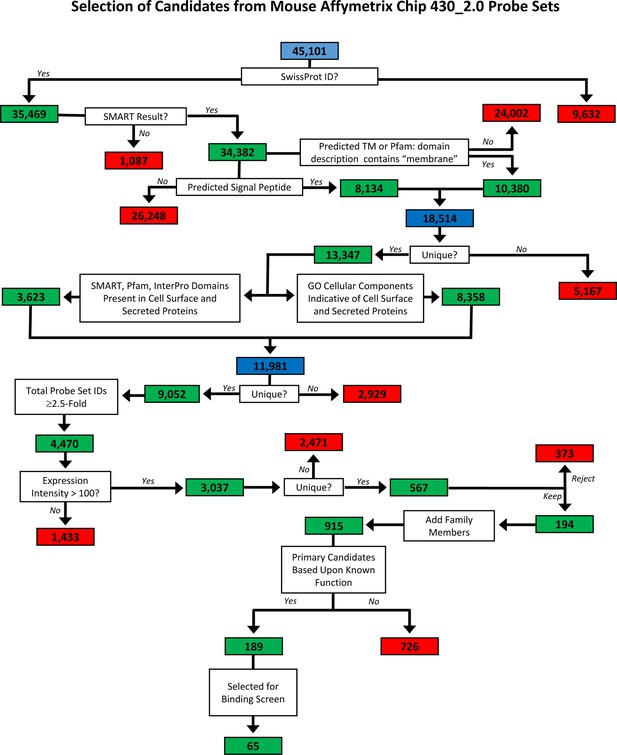
Flow-chart for predicting cell surface and secreted proteins in mouse genome.
The full repertoire of cell surface and secreted proteins encoded in the mouse genome was predicted using a variety of bioinformatics programs as follows. The Mouse Genome 430 2.0 microarray (Affymetrix, CA) contains 45,101 probeset IDs. Of these, 35,469 have UniProtKB/Swiss-Prot identifiers and, as such, correspond to protein-encoding genes. We downloaded the protein sequence for each gene from the UniProtKB/Swiss-Prot database. Protein sequences were submitted to the SignalP server which predicts the presence of a signal peptide (Petersen et al., 2011) and the TMHMM server which predicts the presence of a transmembrane domain (Krogh et al., 2001). Proteins containing a signal peptide and/or a transmembrane domain were analyzed 1) for the presence of domains known to be present in proteins expressed at the cell surface or secreted using SMART (Schultz et al., 1998; Letunic et al., 2012), Pfam (Finn et al., 2014) and InterPro (Hunter et al., 2012) and 2) for gene ontology (GO) cellular component terms consistent with cell surface or secreted proteins (Ashburner et al., 2000). Probeset IDs for genes encoding these proteins were analyzed using dChip software (Li and Hung Wong, 2001) for differential expression amongst the 13 different retinal neuron subtypes. Probeset IDs with ≥3-fold differences in expression amongst the cell subtypes were selected. Genes were ranked according to published data demonstrating that the proteins are known to be involved in cell adhesion, recognition and neuronal guidance or targeting.
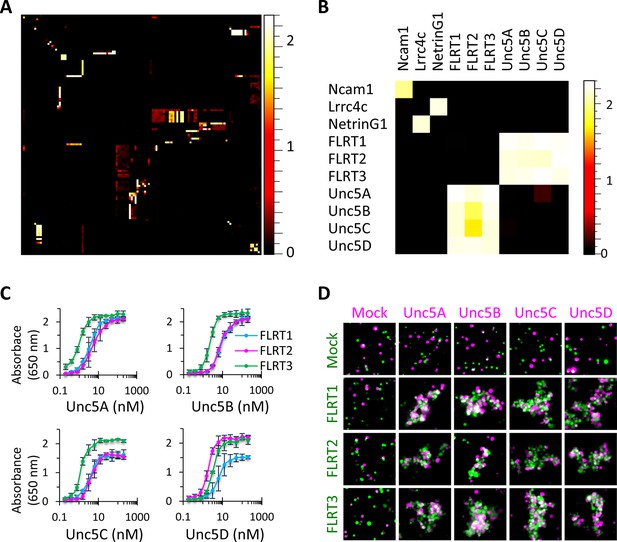
High-throughput binding screen results and FLRT-Unc5 interactions.
(A) 126 x 126 binding matrix. The 126 Fc- and AP-tagged extracellular domain proteins are arrayed along the x and y axes, respectively, in the same order such that homophilic interactions lie on the diagonal. The matrix is colored with a heat map such that high levels of binding are shown in white and no binding is shown in black. Values on the heat map scale represent HRP activity reported as absorbance at 650 nm. Background-subtracted data were deposited in the Dryad database Visser et al., 2015. Western blots of the proteins used in the screen are shown in Figure 2—figure supplement 1 and Figure 2—figure supplement 2. (B) Subset of binding matrix showing FLRT-Unc5 interactions along with Ncam1 homophilic and Lrrc4c-NetrinG1 heterophilic interactions. Heat maps were generated using Image J (Schneider et al., 2012). (C) Titration binding curves to monitor FLRT-Unc5 interactions using purified Unc5 protein binding to FLRT attached to an ELISA plate. FLRT1, blue; FLRT2, magenta; FLRT3, green. Three independent experiments were performed in duplicate and average values are plotted. Error bars represent Standard Deviation. (D) Cell aggregation assays. CHO.K1 cells expressing full length Unc5 (magenta) and FLRT (green) were mixed together and incubated with shaking. Mixed aggregates of magenta and green cells represent trans heterophilic binding. Two independent experiments were performed and representative images are shown.
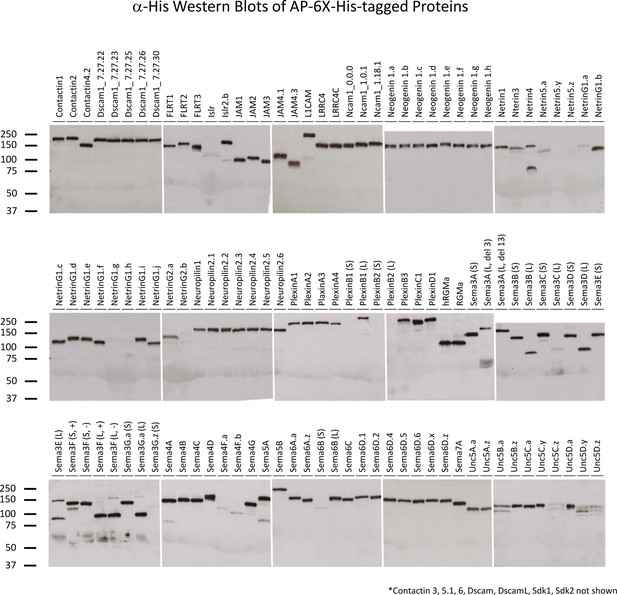
Western blots of proteins for biochemical screen.
α-6X-His Western blots of the AP-6X-His tagged proteins used in biochemical screen were used to assess that recombinant proteins were produced and full-length.

Western blots of proteins for biochemical screen.
α-6X-His Western blots of the Fc-6X-His tagged proteins used in biochemical screen were used to assess that recombinant proteins were produced and full-length.
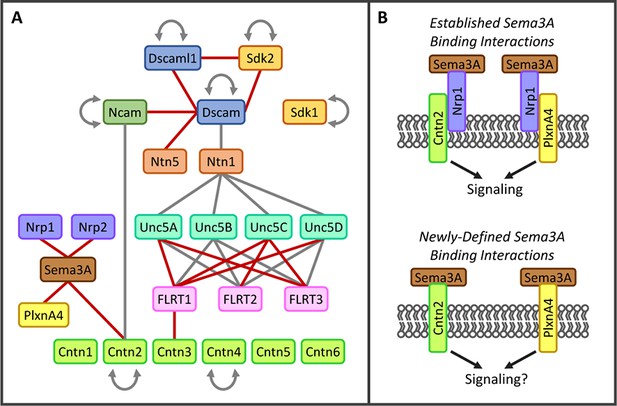
New interactions identified in biochemical screen.
(A) Interactions observed between a subset of proteins included in the screen. Lines indicate direct protein-protein interactions (red line, not previously reported; gray line, previously known). Families of proteins are represented by color. Only one member of the Semaphorin family (Sema3A, brown) and one member of the Plexin family (PlxnA4, yellow) are shown. The complete binding data for all Semaphorins, Plexins and Neuropilins (Nrp, purple) are shown in Figure 4. For space considerations, gene names are used for proteins (e.g. Cntn1 for Contactin1). Figure 1—source data 1 includes full protein names and aliases. (B) (Top panel) Previous studies have demonstrated that Nrp1 (purple) can form a holoreceptor complex for Sema3A ligand (brown) through cis interactions with PlxnA4 (yellow), Cntn2 (green) and a variety of other proteins in the cell membrane (for review see Yazadani and Terman, 2006). (Bottom panel) Our binding screen identified that Sema3A can engage in direct protein-protein interactions with both PlxnA4 and Cntn2 in the absence of Nrp1.
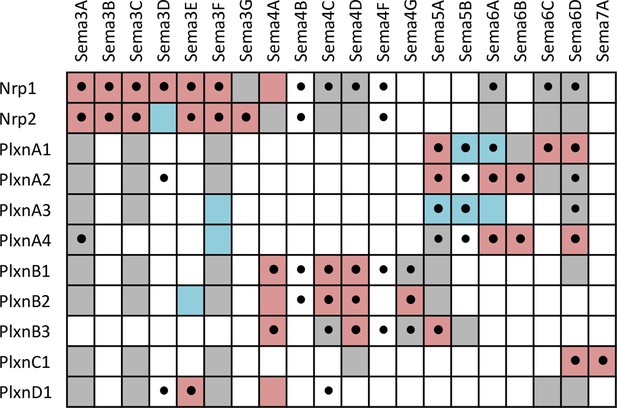
Summary of interactions between Sema-Nrp and Sema-Plxn proteins, highlighting new interactions observed in our screen as well as previously known interactions.
A complete grid of known interactions was compiled from results reported in ten Semaphorin review articles (Yazdani and Terman, 2006; Neufeld and Kessler, 2008; Wannemacher et al., 2011; Hota and Buck, 2012; Neufeld et al., 2012; Yoshida, 2012; Gu and Giraudo, 2013; Roney et al., 2013; Worzfeld and Offermanns, 2014; Masuda and Taniguchi, 2015) and in independent primary literature searches conducted by several members of our laboratory. We included data from ten review articles because there is considerable variability in the interactions reported (see Figure 4—source data 1 and Figure 4—source data 2). All interactions reported in the reviews were corroborated in the primary literature and are denoted in the table by colored boxes that indicate the type of experiment supporting the interaction. Pink = evidence from cell binding assays, surface plasmon resonance, coimmunoprecipitation, transwell suppression and ex vivo explant outgrowth or growth cone collapse. Blue = genetic interactions. Gray, failure to find interaction by one or more of the above methods (i.e. published negative interaction). A black dot (•) indicates a positive interaction observed in our screen. The reference and a description of the supporting data for each previously-known interacting pair are presented in Figure 4—source data 2. It is important to note that there are multiple aliases for most Sema, Plxn and Nrp genes and, as such, our literature searches included these alternative names (e.g. several Sema proteins were initially called collapsins and Sema3B was once called Sema5). These aliases are listed in Figure 4—source data 3.
-
Figure 4—source data 1
Sema-Nrp and Sema-Plxn interactions published in review articles.
A separate binding grid is shown for the interaction pairs reported in each of ten review articles (Yazdani and Terman, 2006; Neufeld and Kessler, 2008; Wannemacher et al., 2011; Hota and Buck, 2012; Neufeld et al., 2012; Yoshida, 2012; Gu and Giraudo, 2013; Roney et al., 2013; Worzfeld and Offermanns, 2014; Masuda and Taniguchi, 2015). Interaction pair boxes are colored in dark gray. The review reference and PubMed ID is listed above each grid. The upper left table with the colored boxes presents a compilation of the interactions reported in all ten review articles. The number in each box represents how many of the ten review articles report the interaction. The boxes are colored using a heat map such that interactions reported by all 10 review articles are colored maroon and those reported by only 1 review article are colored blue. Numbers in yellow font represent interactions that were unverifiable in the primary literature. Unverifiable means that 1) no primary paper was cited for the interaction by the review article and our exhaustive search of the primary literature could not identify a paper reporting the interaction or 2) the interaction was cited by the review article but the paper cited did not test this binding interaction. Note that the unverifiable interactions were reported by only one or two of the ten review artcles (one case, Sema3G-Nrp1,was reported by three out of ten review articles). Unverifiable interactions are determined to be unpublished and are denoted as such in main text Figure 4 but are described in Figure 4—source data 2.
- https://doi.org/10.7554/eLife.08149.012
-
Figure 4—source data 2
Literature search results for Sema-Nrp and Sema-Plexin interactions.
Colored boxes depict interactions reported in ten review articles (Yazdani and Terman, 2006; Neufeld and Kessler, 2008; Wannemacher et al., 2011; Hota and Buck, 2012; Neufeld et al., 2012; Yoshida, 2012; Gu and Giraudo, 2013; Roney et al., 2013; Worzfeld and Offermanns, 2014; Masuda and Taniguchi, 2015). Review-reported interactions that we were able to verify in the primary literature (pink), review-reported interactions that we were unable to verify in the primary literature (yellow; see thorough description in Figure 4—source data 1 legend), reported genetic interactions (blue), reported negative results (gray; yellow font in gray box indicates that this interaction was also reported in one or more review articles but we were unable to verify in the primary literature). A description of the data that determines the color of each box is presented along with the reference for those data (PubMed ID in blue font).
- https://doi.org/10.7554/eLife.08149.013
-
Figure 4—source data 3
Gene name aliases for Sema, Nrp and Plxn.
Aliases were obtained from NCBI Gene and include Mus musculus as well as orthologes in Homo sapiens, Rattus norvegicus, Danio rerio and Gallus gallus. These names were used for conducting primary literature searches to identify published Sema-Plxn and Sema-Nrp interacting pairs.
- https://doi.org/10.7554/eLife.08149.014
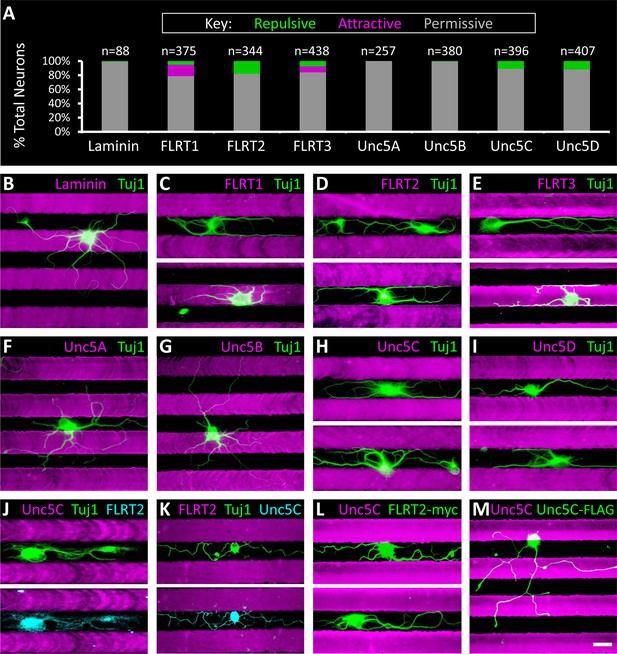
Subpopulations of primary retinal neurons respond to FLRT and Unc5 protein in stripe assays.
Individual retinal neurons harvested from wild- type retinas at P6 were cultured for 4– 6 days on glass coverslips containing alternating stripes of laminin and a purified candidate recognition protein. (A) Quantification showing the percent of neurons that exhibited a repulsive (green), attractive (magenta) or permissive (gray) response to stripes of the candidate recognition protein. n = total number of neurons scored. Raw data are reported in the main text. (B-I) Example images showing responses of neurons to stripes of the indicated FLRT or Unc5 protein (magenta). Stripes were prepared using microfluidic devices as outlined in Figure 5—figure supplement 1 and were visualized by addition of BSA-TRITC (magenta) to the purified FLRT or Unc5 protein patterned. As coverslips were coated with the growth-promoting protein, laminin, prior to application of the stripes, the black (unstriped) regions of the coverslip contain laminin. Neurons were immunostained with an antibody against beta-tubulin (Tuj1; green). (J-K) Example neurons co-stained for Tuj1 (green) and FLRT2 (cyan in J) or Unc5C (cyan in K). Neurons that express FLRT2 are repelled by Unc5C stripes (J), while neurons that express Unc5c are repelled by FLRT2 stripes (K). See main text for quantification. (L-M) Gain-of-function stripe assay. Neurons transfected with full-length FLRT2-myc (green) are repelled by Unc5C stripes (L) whereas, neurons transfected with full-length Unc5C-FLAG (green) are not repelled by Unc5C stripes (M). Scale bar, 30 µm.
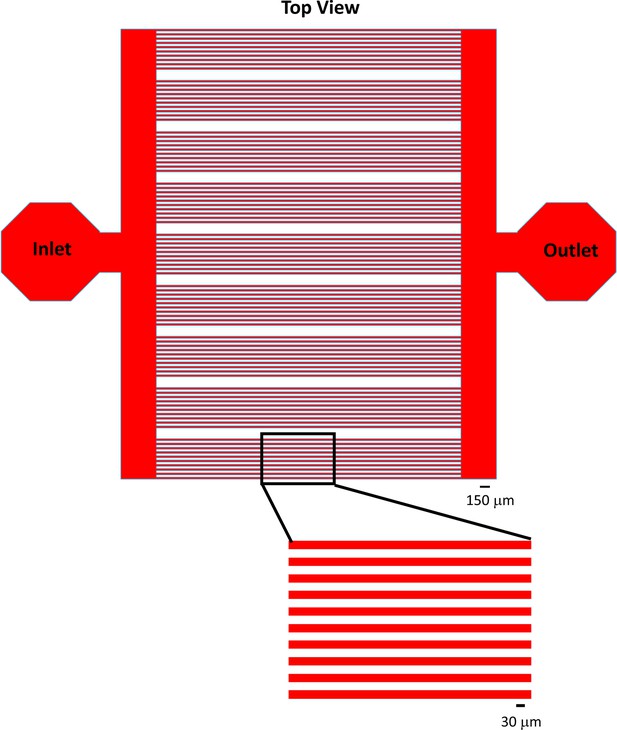
Microfluidic device design for patterning protein stripes for stripe assay.
Top-down view of the microfluidic channels (red) in the PDMS devices. See Materials and methods for additional details regarding channel dimensions and fabrication. Scale bar for upper panel, 150 μm. Scale bar for lower-panel, 30 μm.
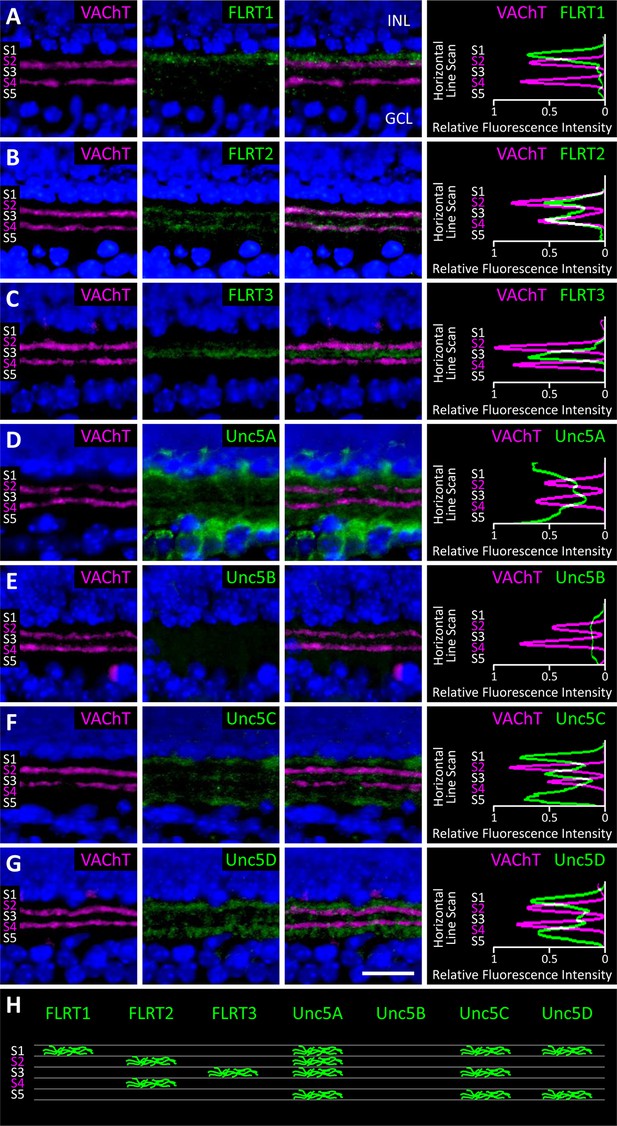
Expression of FLRT and Unc5 proteins in the developing IPL.
(A-G) Retinal sections from C57Bl/6 P6 mice immunostained with an antibody against vesicular acetylcholine transporter (VAChT; magenta), which is expressed by SAC dendrites and thus serves as a marker for sublaminae S2 and S4, and an antibody against one of the FLRTs or Unc5s (green) as indicated in each panel. DAPI (blue) labels cell bodies in the inner nuclear layer (INL) and ganglion cell layer (GCL) flanking the IPL (for schematic see Figure 1B). FLRT and Unc5 antibodies were highly specific as demonstrated by ELISA and shown in Figure 6—figure supplement 1. Expression patterns at P2 and P4 are shown in Figure 6—figure supplement 2. Scale bar, 50 μm. Relative fluorescence of each marker across IPL sublayers S1-S5 is quantified in the histograms plots provided in the right panels. All images were processed together so that the relative fluorescence intensity levels of the staining can be compared amongst different FLRT and Unc5 antibodies. Histogram images produced using ImageJ (Schneider et al., 2012). (H) Schematic summarizing expression pattern of each FLRT and Unc5 protein across IPL sublayers.
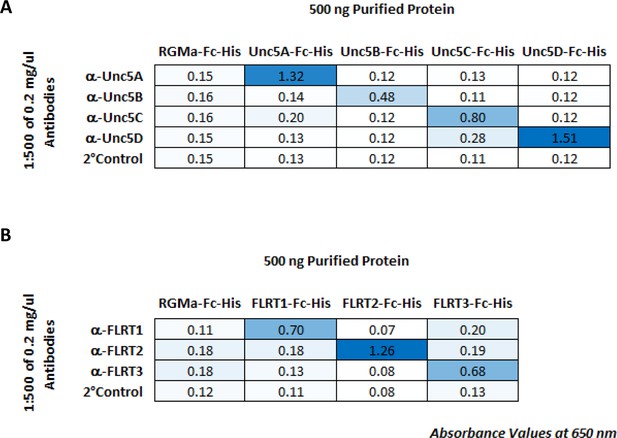
ELISA to test binding specificity of FLRT and Unc5 antibodies.
(A) RGMA-Fc-6X-His (control) and Unc5-Fc-6X-His proteins were captured on a 96-well ELISA plate and stained with each Unc5 antibody in a matrix followed by a secondary antibody conjugated to HRP. Abs650 nm values at 60 min are shown. (B) RGMA-Fc-6X-His (control) and FLRT2-Fc-6X-His proteins were captured on a 96-well ELISA plate and stained with each FLRT2 antibody in a matrix followed by a secondary antibody conjugated to HRP. Abs650 nm values at 60 min are shown.
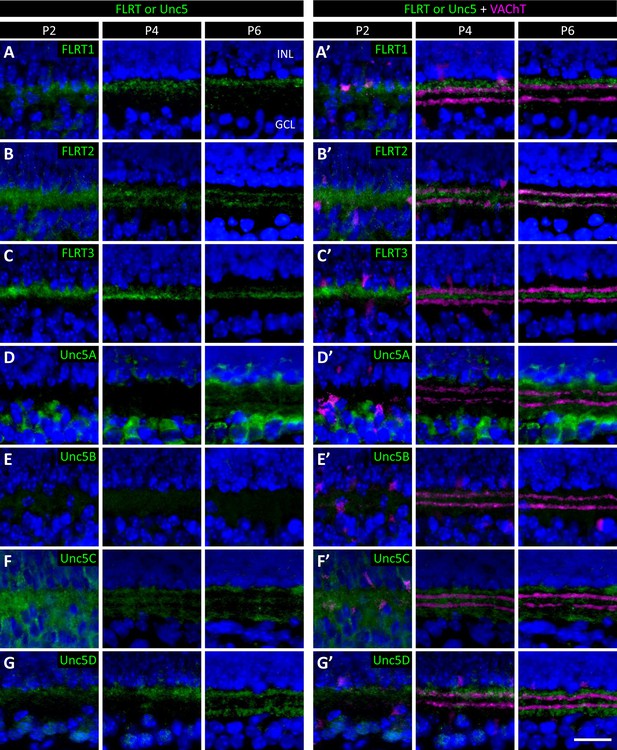
Developmental analysis of FLRT and Unc5 expression in the IPL.
Retinal sections from C57Bl/6 wild type P2, P4 and P6 (P6 images same as Figure 6) immunostained with an antibody against the FLRTs or Unc5s (green) as indicated in each panel. Co-staining of FLRTs and Unc5s with anti-against vesicular acetylcholine transporter (VAChT; magenta), which labels SAC dendrites in sublaminae S2 and S4, is shown in the right panels. DAPI (blue) labels cell bodies in the inner nuclear layer (INL) and ganglion cell layer (GCL) flanking the IPL (for schematic see Figure 1B). Scale bar, 50 μm.
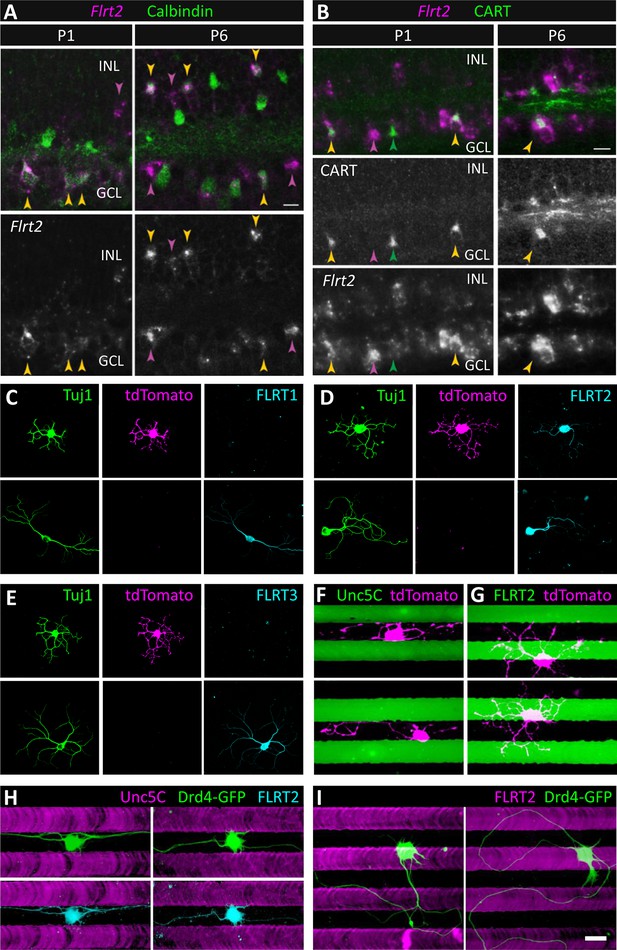
SACs and Drd4-GFP ooDSGCs express FLRT2 and are repelled by Unc5C.
(A) Flrt2 is expressed by SACs, a second amacrine population, and a subset of RGCs. In situ hybridization for Flrt2 RNA (magenta) was combined with immunostaining for calbindin (green), a selective SAC marker at the ages shown (P1 and P6). Yellow arrows indicate Flrt2+ SACs. Cells in the inner nuclear layer (INL) expressing Flrt2 but not calbindin (purple arrows) define a non-SAC Flrt2+ amacrine population. Non-SACs in the ganglion cell layer (GCL) are likely RGCs, based on their large soma size (purple arrows). Among SACs, Flrt2 is detected predominantly in ON SACs (which reside in the GCL) at P1 whereas it is detected more readily in OFF SACs (which reside in the INL) at P6. However, ON SACs positive for Flrt2 are observed at P6 (yellow arrow in GCL), suggesting that Flrt2 is not selective for one SAC population over the other. (B) RGCs expressing Flrt2 include direction-selective ganglion cells (DSGCs). Double staining for Flrt2 and CART, an ooDSGC marker, at P1 and P6. Double-labeled cells (yellow arrows) are observed in the GCL. Not all ooDSGCs express Flrt2, however, as CART+ Flrt2– cells are also apparent (green arrows). Purple arrows indicate Flrt2+ cells that are not ooDSGCs; this group likely includes SACs. Scale bar, 10 µm. (C-E) SACs express FLRT2 protein. Dissociated SACs from P2 Chat-Cre::RosaLSL-tdTomato mice that specifically express tdTomato (magenta) in SACs. Neurons were co-stained with an antibody against Tuj1 (green) and (C) FLRT1, (D) FLRT2, (E) FLRT3 (cyan). Only FLRT2 co-localized with tdTomato-positive SACs. SACs were also negative for Unc5s as shown in Figure 7—figure supplement 1. (F-G) tdTomato SACs (magenta) grown on Unc5C (F) or FLRT2 (G) stripes (green). Stripes were visualized by addition of PLL-FITC to the purified Unc5C or FLRT2 protein patterned. Unc5C (F) but not FLRT2 (G) repelled SACs. (H-I) Dissociated Drd4-GFP ooDSGCs (green) in culture harvested from P3 mice that specifically express GFP in ooDSGCs. (H) Drd4-GFP neurons on Unc5C stripes co-stained with an antibody against Tuj1 (green) and FLRT2 (cyan). (I) Drd4-GFP neurons on FLRT2 stripes stained with an antibody against Tuj1 (green). Neurons cultured 8 DIV. Scale bar, 30 µm.
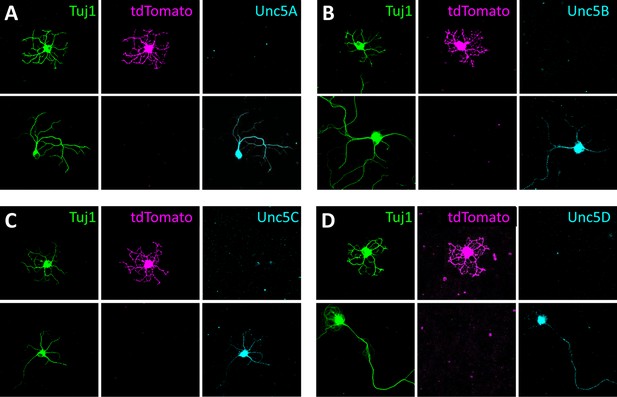
Expression of Unc5s in SACs.
Dissociated SACs (tdTomato, magenta) harvested from P2 Chat-Cre::RosaLSL-tdTomato mice that specifically express tdTomato in SACs. Neurons were co-stained with an antibody against Tuj1 (green) and (A) Unc5A, (B) Unc5B, (C) Unc5C and (D) Unc5D (cyan). None of these co-localized with tdTomato in SACs.





