Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways
Figures
Unwrapping of ssDNA from Escherichia coli SSB under mechanical tension.
(A) Crystal structure (Protein Data Bank ID number 1EYG) and schematic representation of an E. coli SSB tetramer wrapped by 70 nt of ssDNA (blue) in the (SSB)65 mode. From 5′ to 3′, ssDNA interacts with the yellow, purple, green and red subunits. (B) Schematic of SSB unwrapping experiment. A DNA construct consisting of two long double-stranded DNA (dsDNA) handles and a short (dT)70 ssDNA site is tethered between two optically trapped beads in the absence of SSB (Position 1, panel C). When moved to the stream containing SSB (Position 2), a single SSB tetramer binds to the ssDNA site at low tension (∼0.5 pN). The tethered DNA is moved back to the blank stream (Position 1) and a ramping force is applied. Stretching the nucleoprotein complex to >20 pN causes the SSB to dissociate. (C) Experimental flow chamber. Two separate streams containing experimental buffer only (red, Position 1) and buffer plus 0.5 nM SSB (blue, Position 2) form a laminar interface with minimal mixing. (D) Representative force-extension curves (FECs). Relaxing curves (red) are obtained after SSB dissociation, and are well fit to a polymer model of bare DNA (black dotted line, ‘Materials and methods’). Stretching curves (purple) of the SSB-ssDNA complex deviate from a model assuming the protein adopts the (SSB)65 wrapping mode at all forces (black dashed line). Cartoon illustration of SSB unwrapping shows the SSB behavior at particular forces. (E) Change in extension upon SSB wrapping vs applied force. The change in extension is determined from the extension difference between stretching and relaxing curves in (D). Individual traces (gray) are binned and averaged to yield a mean change in extension (black opened circle; error bars are S.D.). The data deviates from the model (dashed line, determined from the difference between the dashed and dotted lines in (D)) at forces >1 pN. Representative traces (red, green, and blue) display the differences between the individual and averaged traces.
Dissociation of SSB upon DNA stretching.
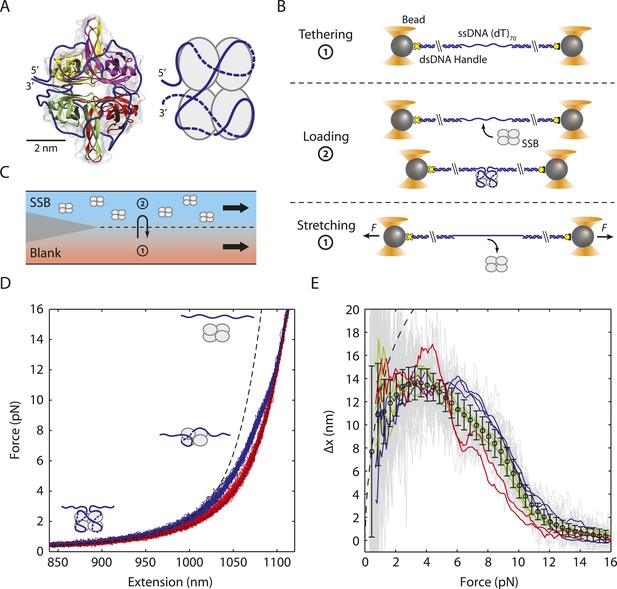
Averaged stretching (blue) and relaxing (red) FEC from Figure 1D, and bare DNA FEC (green). Both the relaxing and bare DNA stretching curves are fitted to the polymer elasticity model with 3260 bp dsDNA handles and 70 nt ssDNA (black dashed line, ‘Materials and methods’). The model assumes zero extension at zero force and fits the data. The resulting fits are consistent with each other, indicating that SSB has dissociated during stretching. Error bars are S.D.
Single-stranded DNA polymer modeling.
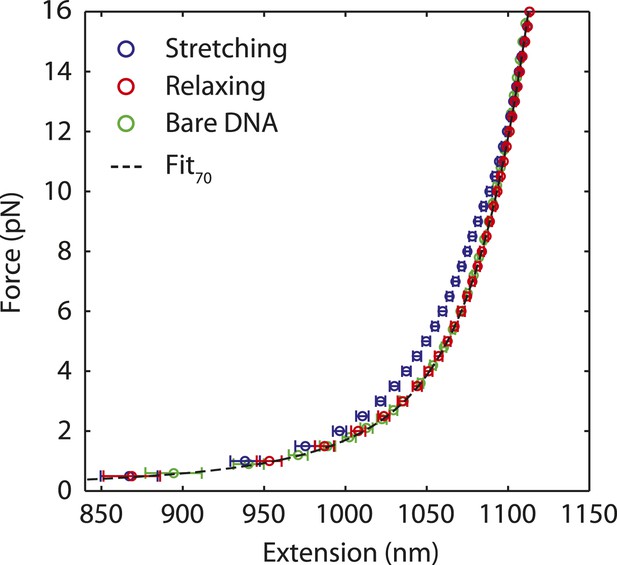
Representative FEC of stretching and relaxing a DNA construct containing 3260 bp dsDNA handles and 70 nt (green) or 140 nt (orange) ssDNA. The total extension of the tether is modeled by the sum of dsDNA and ssDNA extensions. The dsDNA segment is modeled using the extensible worm-liked chain (XWLC), while the ssDNA segment is fitted to the snake-like chain (SLC; ‘Materials and methods’). Black dashed and dotted lines are fits to the 70 nt and 140 nt ssDNA constructs, respectively. The extension difference (inset, blue) between 70 nt and 140 nt ssDNA constructs illustrates the validity of the ssDNA elasticity model over short lengths (70 nt).
Dissociation force of SSB-ssDNA.
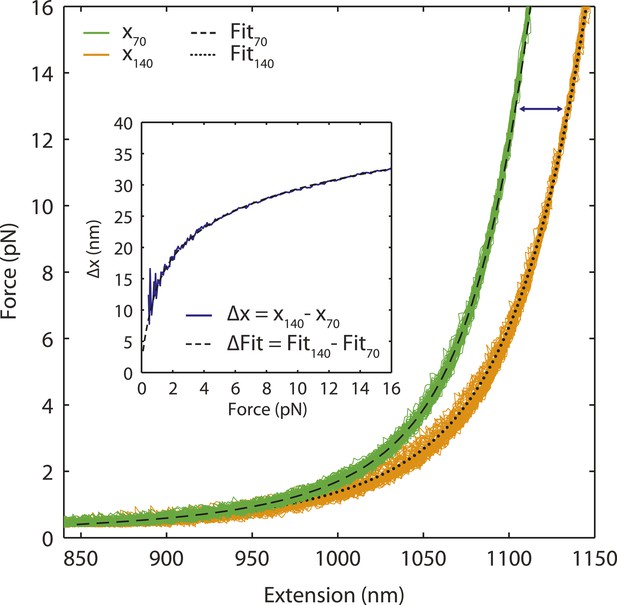
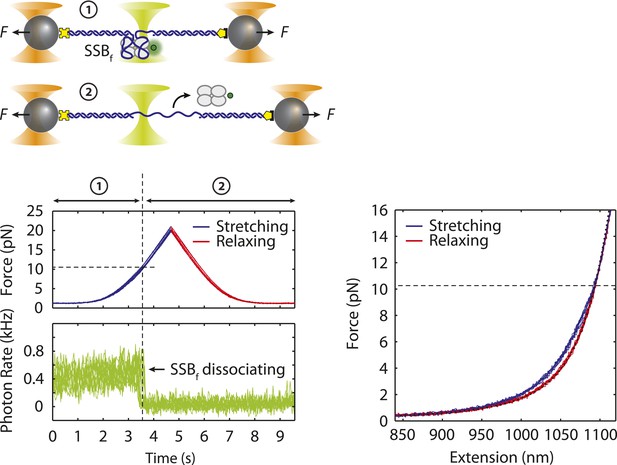
Cartoon schematic and representative traces showing combined fluorescence and DNA extension measurements. A DNA construct bound by fluorescently labeled SSB, SSBf, is stretched (blue) and relaxed (red) under mechanical force. Upon reaching a force ∼10 pN, SSBf dissociates from the DNA as indicated by the decrease in fluorescence. The relaxing curves from the corresponding FECs match the polymer elasticity model of bare DNA (black dotted line, ‘Materials and methods’) indicating that the SSB has dissociated during stretching. The dissociation force from the FECs is consistent with the fluorescence data.
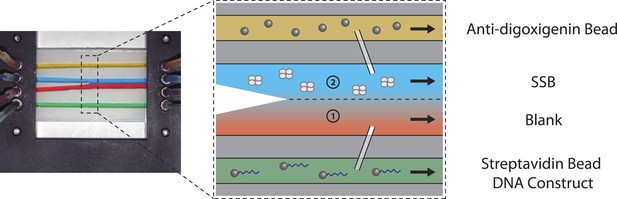
Sample chamber.
Image and schematic of a laminar flow chamber. Two glass coverslips are used to sandwich patterned parafilm (Nescofilm). For illustration purposes, food dye of different colors is flowed into the chamber via inlet tubing at a rate of 100 µl/hr. Two streams, one containing experimental buffer only (red, 1), and the other containing buffer plus SSB (blue, 2), merge into the central channel but do not mix appreciably due to the laminar flow. The chamber design allows rapid exchange of buffer conditions by moving the optical traps across the stream interface. The top channel (yellow) is loaded with anti-digoxigenin beads, while the bottom channel (green) is loaded with DNA-bound streptavidin beads. Both beads diffuse through glass capillaries into the middle channel where the optical trapping experiment is performed.
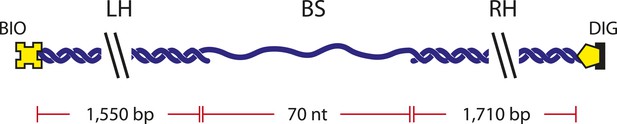
DNA construct.
Schematic of single-stranded DNA construct. The DNA construct consists of three separate fragments ligated together (‘Materials and methods’): ‘Right Handle’ (RH), ‘Left Handle’ (LH), and ‘Binding Site’ (BS). The handles served as functionalized linkers that connect to trapped beads through biotin-streptavidin and digoxigenin-anti-digoxigenin linkages and spatially separate the beads from the protein binding site.
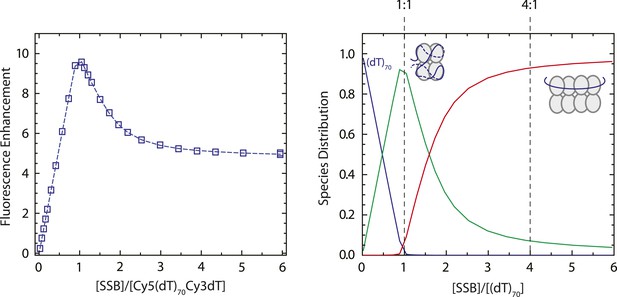
SSB binds to dT70 in the fully wrapped (SSB)65 mode at a 1:1 molar ratio in 100 mM Tris buffer.
Results of an equilibrium titration of Cy5-(dT)70-Cy3-dT-3′ (0.1 μM) with SSB (left panel; 100 mM Tris-HCl, 20 mM NaCl, 0.1 mM EDTA, 25°C) plotted as normalized Cy5 fluorescence (Fn = (F − F0)/F0) vs molar ratio of total SSB protein (tetramer) to total DNA concentrations (where F0 is the fluorescence intensity of DNA alone and F is the fluorescence measured at each point in the titration). The biphasic character of the binding isotherm indicates that two types of complexes can form, the first having one and the second having two tetramers bound and characterized by high and intermediate FRET values ((SSB)65 and (SSB)35 modes, respectively). The continuous line represents the best fit to the data based on a two-site model (Roy et al., 2009) with equilibrium binding constants, k1 = 1 × 1010 M−1 (minimum estimate) and k2 = (1.21 ± 0.04) × 108 M−1 and two additional parameters F1 = 10.1 ± 0.1 and F2 = 4.8 ± 0.1, reflecting the maximum Cy5 fluorescence observed for one and two tetramers bound, respectively. Species distribution predicted from the best fit parameters listed above (right panel). At low concentration of SSB tetramers the protein binds to dT70 exclusively in the fully wrapped (SSB)65 binding mode, although as the SSB concentration increases ([SSB]tot/[dT70]tot > 1) the (SSB)35 binding mode starts to form in which two SSB tetramers are bound to one molecule of dT70.
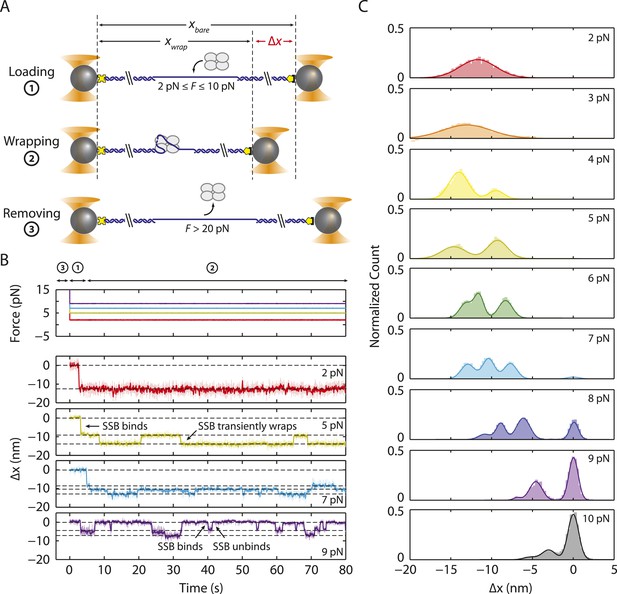
Intermediate ssDNA wrapping states of SSB under tension.
(A) Schematic of SSB constant force wrapping experiment. A DNA construct is held between two optical traps under a constant tension between 2–10 pN in the presence of protein. An extension change, Δx, is measured upon SSB binding, wrapping or unwrapping ssDNA. At the end of each observation, SSB is removed by stretching the DNA construct to high force (>20 pN). (B) Representative time traces of SSB-ssDNA wrapping at 2, 5, 7, and 9 pN (red, green, blue, and purple respectively). Extension change data were acquired at 66 kHz and boxcar averaged to 10 Hz (dark color). In all traces, SSB first binds and compacts ssDNA as indicated by an extension decrease. Depending on tension, SSB displays several intermediate wrapping states. Black dashed lines represent the mean extension change of each particular wrapping state. (C) Extension change distribution from many SSB wrapping traces at constant tensions between 2–10 pN. The color map matches that in (B). Solid lines are multi-Gaussian fits to the distributions.
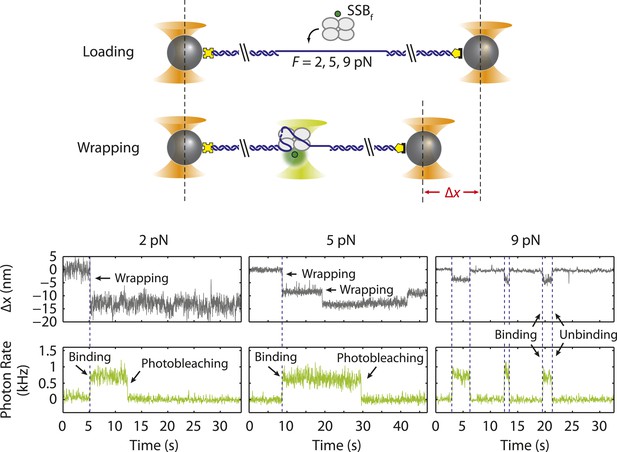
Single SSB binding and wrapping transitions.
Schematic and representative traces illustrating a wrapping experiment with fluorescently labeled SSB, SSBf. A DNA construct is held between two optical traps at a constant tension of 2, 5, and 9 pN (left, middle, and right panels). An extension change, Δx, is measured upon SSBf wrapping or unwrapping ssDNA. Upon SSBf binding, a decrease in extension (gray) and increase in fluorescence (green) are observed simultaneously (all panels). A further decrease in extension (middle panel) does not result in further increase in fluorescence, indicating that the same SSB wraps additional ssDNA. At high forces (right panel) extension increases correspond to SSB dissociation.
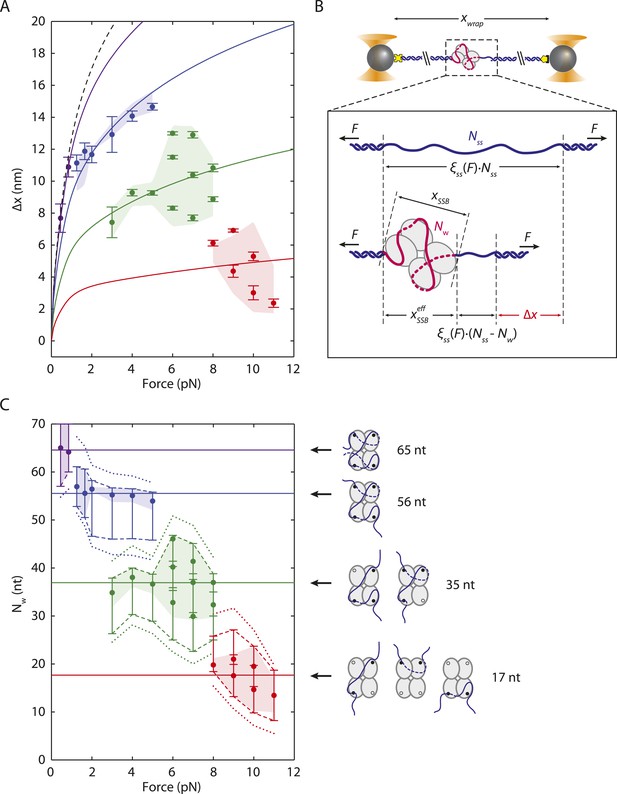
SSB wrapping modes.
(A) Mean change in extension Δx vs tension for each wrapping state, derived from the peaks of the distributions in Figure 2C. Error bars represent S.E.M. and were determined by bootstrapping. The dashed line is the model in Figure 1D. Solid lines represent models of Δx based on Equation 3 for SSB wrapping Nw = 65, 56, 35, and ∼17 nt (purple, blue, green, and red, respectively; ‘Materials and methods’). Data points are clustered into 4 groups corresponding to those states (purple, blue, green, and red circles). (B) Schematic representation of Δx. Top: Bare ssDNA (with Nss = 70 nt) and its extension, xbare, based on a polymer elasticity model Equation 1 (‘Materials and methods’). Bottom: SSB-wrapped ssDNA showing the number of wrapped nucleotides, Nw (<70, red) and the remaining unwrapped nucleotides (Nss − Nw, blue). The extension of wrapped DNA, xwrap is calculated from an elasticity model and the effective physical size of the SSB-ssDNA complex, , Equation 2 (‘Materials and methods’). Δx is the difference between xwrap and xbare, Equation 3. (C) Number of wrapped nucleotides Nw vs tension F. Each data point in (A) is mapped to Nw using the model described in the text (‘Materials and methods’; Figure 3—figure supplement 1). Dotted lines represent the maximum possible range of Nw for each colored group of points based on being <6.5 nm (Figure 3—figure supplement 1, left panel). Dashed lines represent a tighter range of possible Nw for each group of points derived from the SSB-ssDNA structure (Figure 3—figure supplement 1, middle panel). Error bars represent this range for each individual data point. The shaded areas represent the tightest range of possible Nw for each group based on the ‘hotspot’ analysis described in the text (Figure 3—figure supplement 1, right panel). The points are the best estimates of Nw from the model. The shaded areas and solid lines in (C) map directly to those in (A). Cartoon schematics depict possible wrapping modes corresponding to the 4 groups.
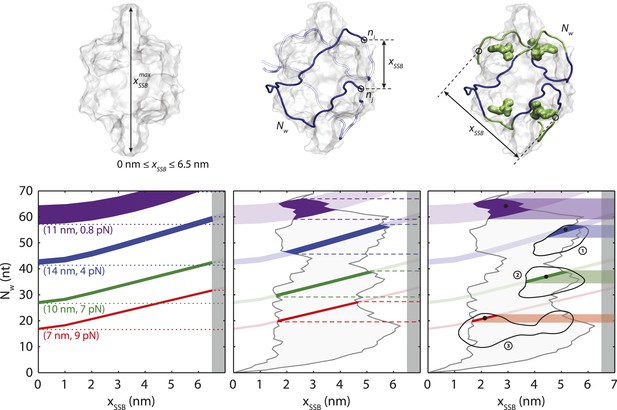
SSB wrapping models.
Three-level modeling of SSB wrapping configurations. Schematics of SSB, wrapped ssDNA (blue), and the distance between wrapped ends, xSSB (black arrow; top panels). Each extension change data point Δx(F) in Figure 3A corresponds to a curve in the space of possible Nw and xSSB, according to Equation 5 (colored curves, bottom panels). The widths of the curves correspond to the error bars in Figure 3A. Selected data points from Figure 3A are displayed (purple: F = 0.8 pN, Δx = 11 nm, blue: 4 pN, 14 nm, green: 7 pN, 10 nm, and red: 9 pN, 7 nm). At the first level of modeling (left panels), xSSB is assumed to be limited only by the size of the protein (i.e., xSSB < 6.5 nm; dark gray shaded area). The range of possible Nw corresponding to each selected data point is shown by the colored dotted lines. At the second level (middle), the range of possible xSSB is refined by utilizing the (SSB)65 crystal structure. The end-to-end distance between every pair of nucleotides ni and nj along the ssDNA in the structural model defines a lower and upper bound of xSSB for each Nw (gray shaded area). This, in turn, narrows down the range of possible Nw for each data point (colored dashed lines). At the third level (right), four ‘hotspots’, residues on each SSB monomer with which nucleotides interact most strongly (green molecular surfaces in the schematic and green nucleotides), are used to refine the estimates for xSSB further. Three regions near the hotspots (black contours) are identified and used to calculate xSSB. The numbering (1, 2, and 3) corresponds to the configurations shown in Figure 3—figure supplement 2. This analysis provides the narrowest estimate for the range of Nw for each data point Δx (colored bands). The best estimates for Nw are obtained from the center of this range (black dots); these are plotted in Figure 3C vs force.
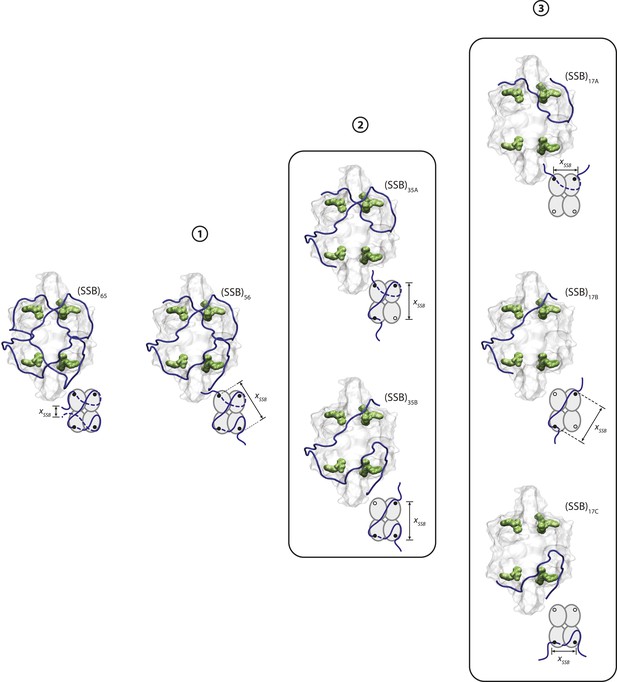
SSB wrapping pathway.
Crystal structures and schematics of SSB wrapping ssDNA (blue) in different wrapping modes. Each mode illustrates possible wrapping configurations that correspond to the regions, numbered 1, 2, and 3 in Figure 3—figure supplement 1. As tension increases (from left to right), SSB wraps less ssDNA, and the number of hotspots interacting with ssDNA (green molecular surfaces in structures, black dots in schematics) decreases.
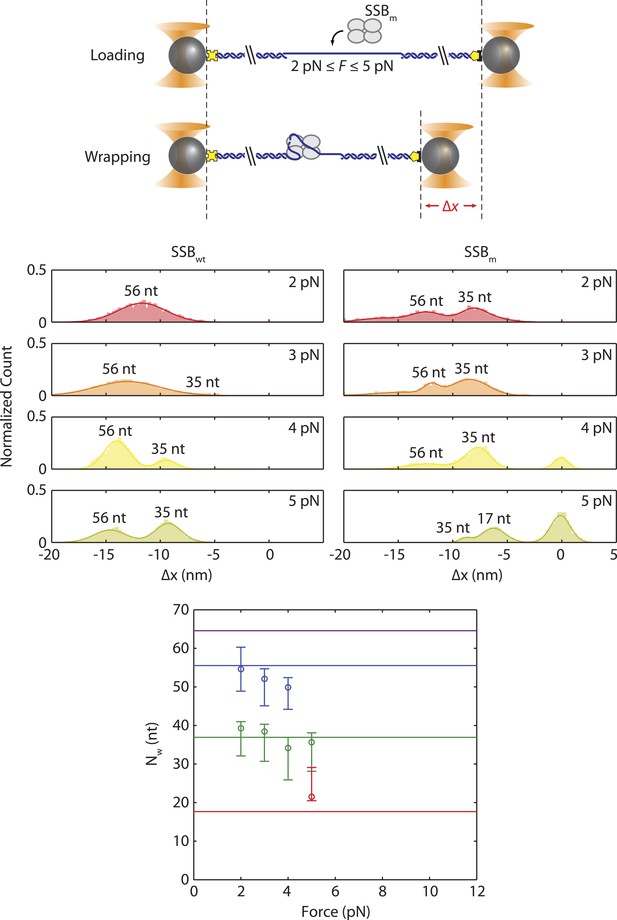
Wrapping modes of SSB mutant.
Schematic of wrapping experiment using SSBm, a SSB mutant in which Trp-54 is replaced by Ser-54. Comparison of extension change distributions between wild-type SSB (left panels) and SSBm (right). At the same tensions (3–5 pN), SSBm wraps less ssDNA than wild-type SSB, and is more likely to wrap 35 nt. The mean number of wrapped nucleotides vs tension was estimated in the same way as for wt SSB (Figure 3C).
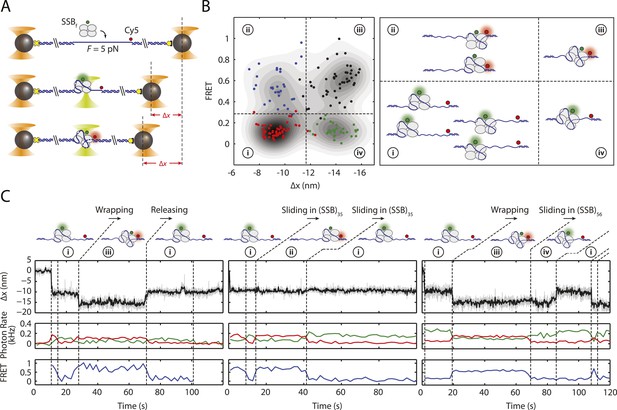
SSB binding modes and diffusion mechanism.
(A) Schematic of fluorescently labeled SSB, SSBf, ssDNA wrapping experiment. A Cy5-labeled DNA construct is tethered between two optical traps under a constant tension of 5 pN. Upon binding of an AlexaFluor555-labeled SSB, both DNA extension change, Δx, and single-molecule FRET are measured simultaneously. (B) Scatter plot of FRET efficiency and Δx. Data (circles) are assigned to 4 states (red (i), blue (ii), black (iii), and green (iv)) based on the value of FRET and Δx. A density map of the combined FRET-extension states overlaid with the scatter plot confirms that the data can be separated into 4 states. Cartoon illustrations of nucleoprotein complexes demonstrate possible SSB wrapping configurations corresponding to the 4 assigned states. (C) Representative traces showing combined fluorescence and DNA extension measurements. Change in extension (top; boxcar averaged to 50 Hz) and fluorescence (middle; boxcar averaged to 0.5 Hz) of donor (SSBf, green) and acceptor (Cy5, red) are measured simultaneously. Together, FRET efficiency (bottom; blue) and extension change (top; black) reveal the SSB wrapping states (i and ii, iii and iv) and their dynamics (ssDNA wrapping/releasing and sliding).
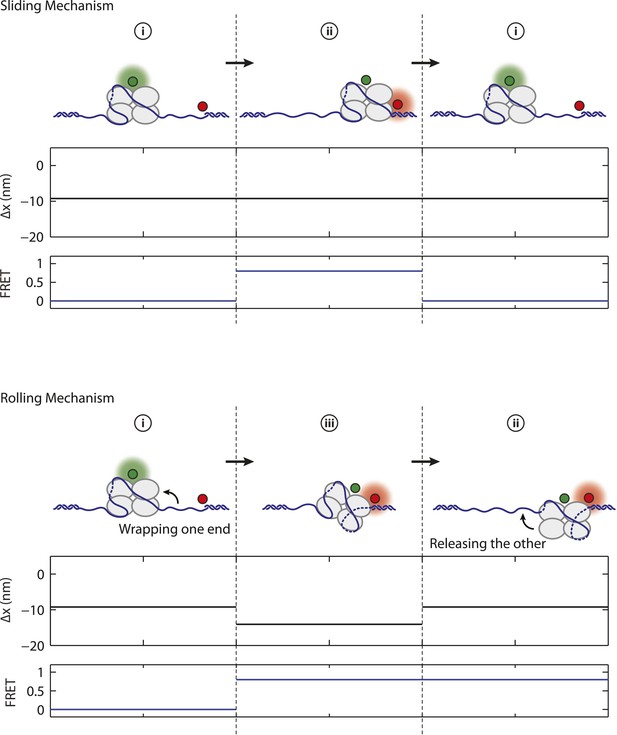
Mechanism of SSB diffusion.
Cartoon illustrations of nucleoprotein complexes diffusing along ssDNA with different proposed mechanisms. Schematic FRET efficiency and Δx displaying multiple transitions between states (i, ii, iii, iv). In a sliding or reptation mechanism, FRET transitions occur independently of changes in wrapping state (top panel). A rolling mechanism involves SSB displacement by wrapping one end of DNA followed by releasing the other (bottom panel; i → iii → ii or ii → iii → i). No examples (0 of N = 82) of rolling are observed in our experiment.
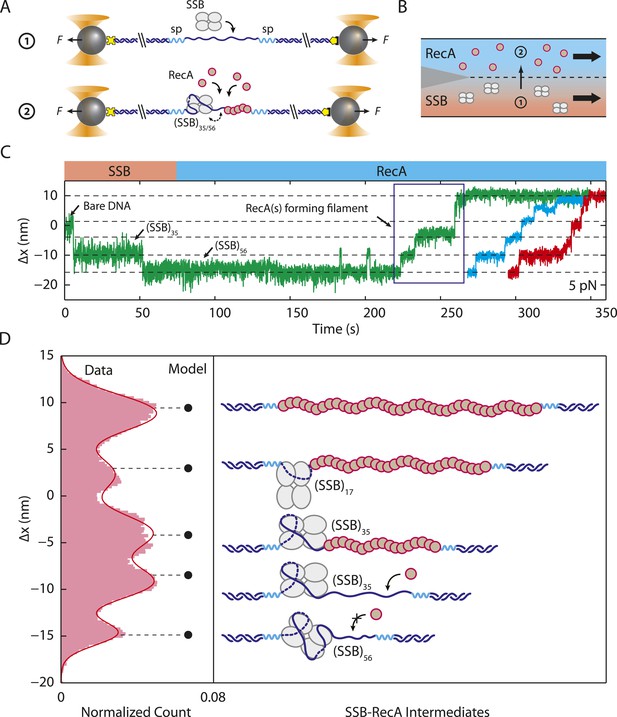
Unwrapping of ssDNA from SSB by RecA filament formation.
(A) Schematic representation of SSB-RecA experiment. A standard DNA construct consisting of a 70-nt single-stranded DNA ((dT)70) fragment was synthesized to contain two internal 18-atom hexa-ethylene-glycol spacers at both ss-dsDNA junctions (cyan; ‘Materials and methods’). The spacers prevent RecA filament formation onto the dsDNA. The construct is tethered in the presence of SSB (Position 1). After the SSB binds, the tethered DNA is moved to the stream containing RecA for observation (Position 2). (B) Experimental flow chamber for SSB-RecA experiment. Two separate streams contain experimental buffer plus 0.5 nM SSB (red, Position 1) and buffer plus 125 nM RecA and 125 μM ATP-γS (blue, Position 2). (C) Representative time traces showing competition between RecA and SSB on ssDNA (green, blue, red). Transient wrapping-unwrapping of SSB slows down the nucleation of RecA. Formation of RecA filament extends ssDNA (blue box), displaces the SSB, and stops after reaching the spacers at the ss-dsDNA junctions. The dotted lines correspond to the model in (D). (D) Extension change distribution of SSB-RecA intermediates at a constant tension of 5 pN (pink) obtained from many RecA filament formation time traces (N = 25). Five states representing SSB-RecA dissociation intermediates are illustrated (schematics) and assigned to peaks of the distribution. Extensions corresponding to these states are predicted using polymer models of elasticity (black dots and dotted lines, ‘Materials and methods’).
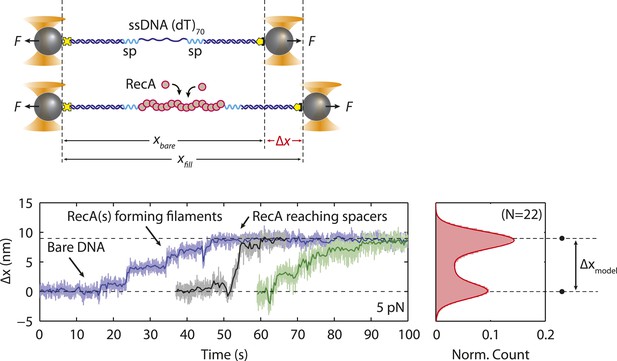
RecA filament formation on modified single-stranded DNA.
Schematics and representative time traces showing RecA filament formation experiment. A DNA construct consisting of two long dsDNA handles, a short 70-nt ssDNA site, and two spacers (cyan, ‘Materials and methods’) is held between two optical traps at a constant tension of 5 pN in the blank buffer. The construct is then moved into the buffer stream containing 125 nM RecA and 125 µM ATP-γS. A change in extension, Δx, is measured while RecA polymerizes, extending the ssDNA. Upon reaching the spacers, RecA filament formation stalls. The extension change distribution from many RecA filament formation time traces (blue, black, green; N = 22) are consistent with the polymer elasticity model of bare DNA and RecA-filled DNA (black dots; ‘Materials and methods’), indicating that RecA has fully polymerized on ssDNA.
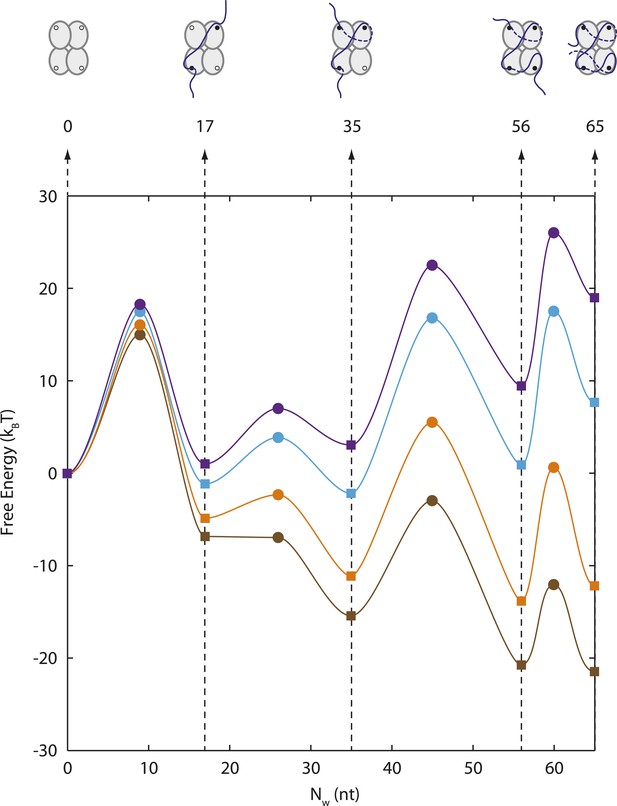
Energy landscape of SSB wrapping.
Energy landscapes of a single SSB wrapping ssDNA at representative forces reconstructed from extension change probability distributions vs tension (Figure 2C). The potential wells correspond to the stable SSB-ssDNA intermediates (cartoon schematics): (SSB)65, (SSB)56, (SSB)35, (SSB)17, and unbound, respectively. The energy associated with each intermediate is determined from the occurrence probabilities for each state (squares, ‘Materials and methods’). The barrier heights and positions (circles) are determined from the state lifetimes (‘Materials and methods’). In the absence of tension, SSB wraps ssDNA in the (SSB)65 binding mode. Increasing tension (brown, orange, cyan, purple lines correspond to 0, 3, 7, 9 pN, respectively) tilts the energy landscape, changes the free-energy difference between wrapping intermediates, and favors different SSB-ssDNA binding modes.
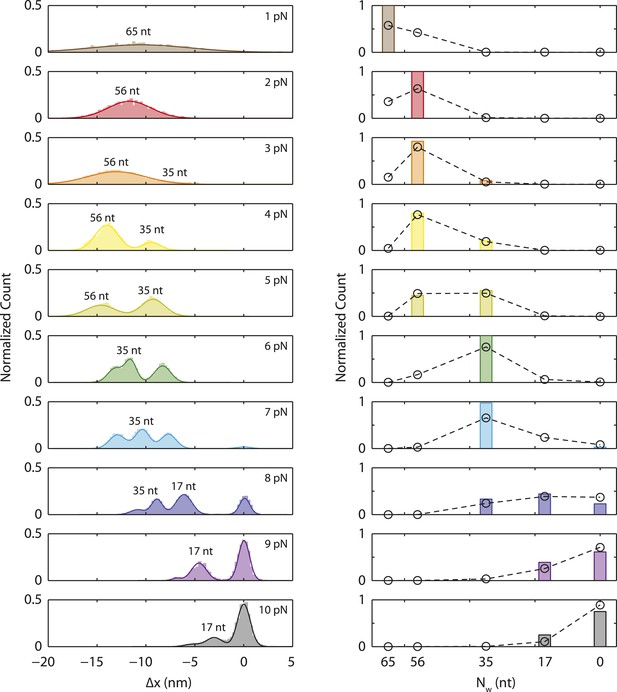
Occurrence probability of SSB wrapping intermediates.
Extension change distributions (left panels) of many SSB wrapping events obtained from force-ramp experiments (1 pN) and constant force experiments (2–10 pN). Individual wrapping intermediates are analyzed and assigned to corresponding SSB binding modes based on Figure 3C. At all tensions, the probability of each SSB binding modes (right panels, color bars) is derived from the area under the distributions. The model (black circles, ‘Materials and methods’) obtained from the energy landscape in Figure 6 matches well with the experimentally derived probabilities.
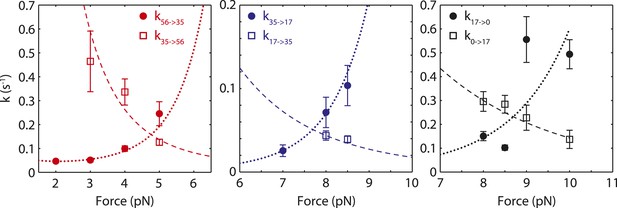
Modeling of transition rates between SSB wrapping intermediates.
Unwrapping (solid circles) and wrapping (open squares) transition rates between different SSB wrapping intermediates vs force. The rates were determined from dwell times and transition probabilities in Figure 2C (‘Materials and methods’). The data were fit globally (unwrapping, dashed line; wrapping, dotted line) using expressions of the form Equation 11 and Equation 12 using as parameters the three barriers and distances to the transition state , , , , , and (‘Materials and methods’).





