A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division
Figures
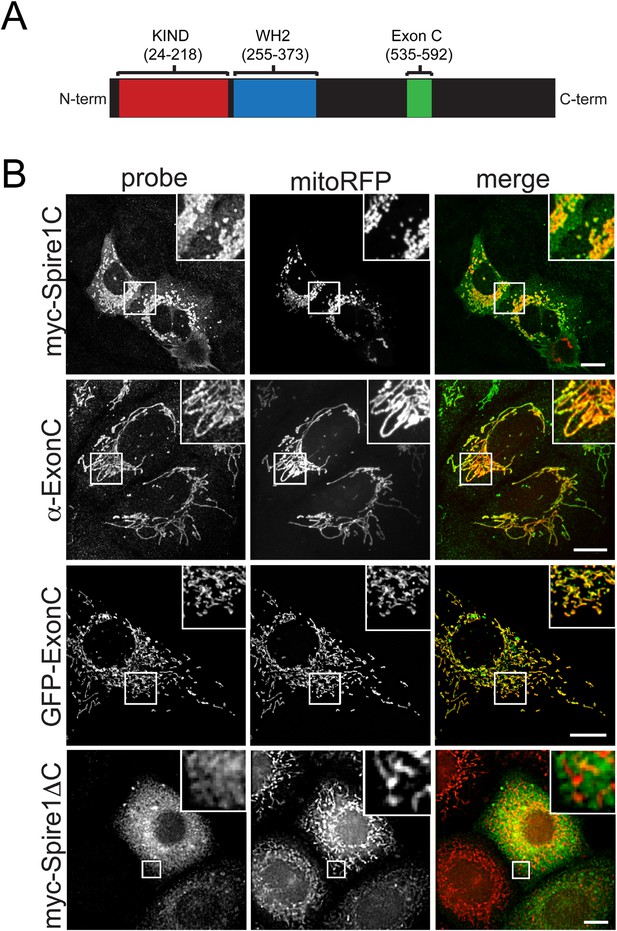
The Spire1 alternate exon ExonC is necessary and sufficient for localization to mitochondria.
(A) Full length Spire1C domain structure: The number ranges indicate the amino acid regions of the conserved domains probed in this study. (B) Spire1C localizes to mitochondria. myc-Spire1C: U2OS cells cotransfected with myc-Spire1C and mitoRFP show robust localization of myc-Spire1C to mitochondria. α-ExonC: U2OS cells stained with an antibody raised against ExonC (α-ExonC) and expressing mitoRFP show endogenous Spire1C labeling on mitochondria. GFP-ExonC: U2OS cells cotransfected with GFP-ExonC and mitoRFP show robust targeting of GFP-ExonC to mitochondria. myc-Spire1ΔC: U2OS cells cotransfected with myc-Spire1ΔC and mitoRFP show no specific targeting of myc-Spire1ΔC to mitochondria. All cells were fixed and primary antibodies were counterstained with Alexa-488 secondary antibody before imaging with confocal fluorescence microscopy. Scale bars: 10 μm. Inserts are magnifications of the boxed regions.
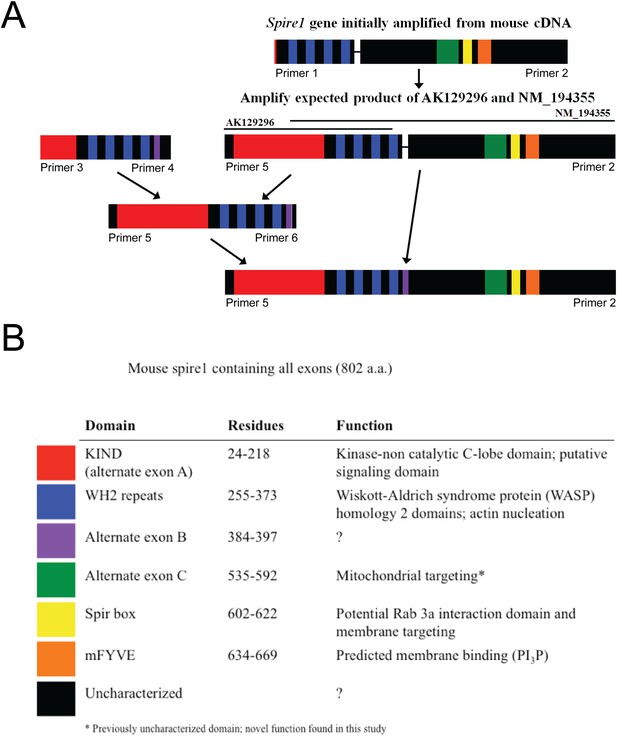
Construction of the complete Spire1C protein sequence as explained in detail in the ‘Materials and methods’.
Spire1C gene amplified from mouse brain cDNA. (A) Initial construct based on NM_194355 was later combined with the sequence based on AK129296 to form full-length Spire1C protein of 802 amino acids with all known exons. (B) Spire1C diagram showing both characterized (red, blue, yellow, and orange) and uncharacterized (purple, green, and black) domains (indicated in the legend on the bottom).
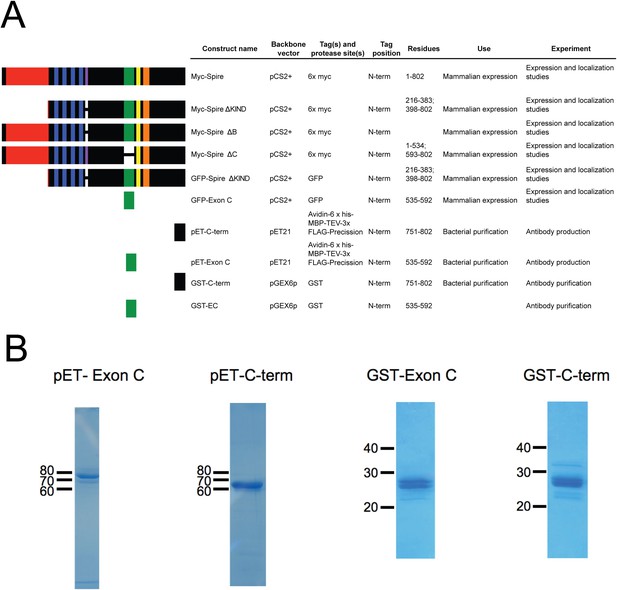
Constructs used to probe Spire1 function.
(A) Spire1C constructs used in this study. Residue numbers are for the 802 a.a. full-length mouse Spire1C protein. (B) Coomassie-stained gels of purified proteins used for antibody production (pET constructs) and affinity purification (GST constructs).
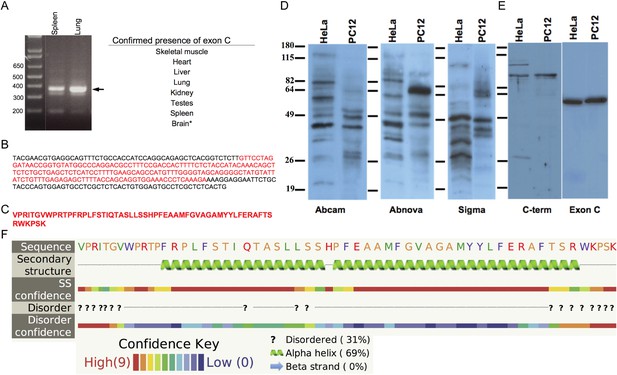
Spire1C contains a previously uncharacterized alternate exon of 58 amino acids.
(A) Spire1 ExonC was found to be present in the tissues listed. *PCR from cDNA did not yield conclusive results, but the ExonC-containing gene was originally amplified from mouse brain cDNA. (B) Sequencing results from TOPO pCR2.1 vector using the M13 reverse primer. ExonC sequence is shown in red. (C) Translated protein sequence from red portion in C. (D) Representative immunoblots comparing commercial antibodies to Spire1 used to detect endogenous Spire1 in HeLa and PC12 cell lysates, 25 μg per lane. (E) Affinity purified Spire1 C-terminal antibody and ExonC antisera (both generated in this study) tested on HeLa and PC12 cell lysates, as in (A). Strikingly different protein bands are observed for the same cell type with different antibodies and between cell types with the same antibody. Molecular weight protein markers are the same size for each blot, though the positions vary slightly (as indicated). (F) The Phyre server was used to predict any secondary structural elements in ExonC. Phyre compiles structure predictions from three different algorithms (psipred, jnet, and sspro) to form a consensus prediction and probability for each structural element. Probability ranges from 1–10, with 10 being the most probable. Yellow amino acids are small/polar, green are hydrophobic, red are charged, and purple are aromatic + cysteine.
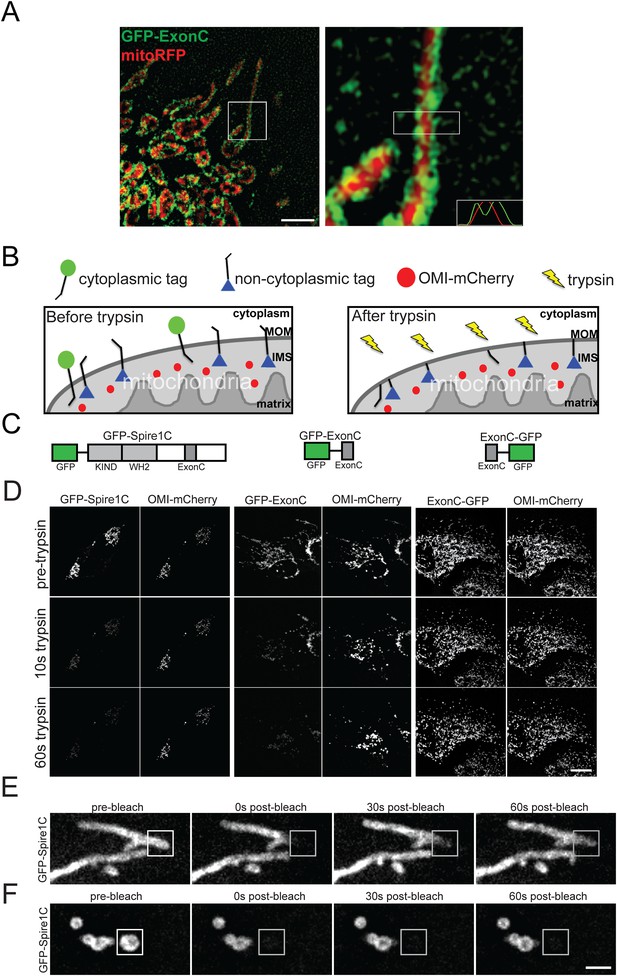
Spire1C localizes to the mitochondrial outer membrane with its formin and actin binding domains facing the cytoplasm.
(A) ExonC localizes to the peripheral region of mitochondria. Left: structured iIllumination microscopy (SIM) image of a U2OS cell transfected with GFP-ExonC and mitoRFP reveals localization of GFP-ExonC to the periphery of mitochondria. Right: Magnification of boxed region on left. The insert on the lower right corner is a fluorescence intensity linescan of the rectangular boxed region indicating the inversely related profiles of GFP-ExonC vs mitoRFP. Scale bar: 10 μm. (B) Illustration of the principle of the fluorescence protease protection (FPP) assay performed on mitochondrial outer membrane (MOM) proteins. If a fluorescent protein tag faces the cytoplasm (green circle), it is degraded by trypsin and its fluorescence is depleted. If the protein tag faces the interior of the mitochondria (blue triangle), it is protected from trypsin in the cytoplasm, and thus its fluorescence remains after trypsin addition. An intermembrane space (IMS) marker, OMI-mCherry, serves as a control to verify that the mitochondrial outer membrane has not been permeabilized by the digitonin treatment, and furthermore to confirm that trypsin is not degrading proteins in the IMS. (C) Schematic of the constructs used in our FPP assays. (D) Cells cotransfected with OMI-mCherry and GFP-Spire1C (N-terminal GFP tag, left) or GFP-ExonC (N-terminal GFP-tag, middle) were treated with 20 μM digitonin and 4 mM trypsin. In both cases OMI-mCherry fluorescence remained, whereas the N-terminal GFP tags were mostly depleted within 60 s after trypsin treatment. In contrast, in cells transfected with ExonC-GFP (C-terminus GFP tag), GFP fluorescence remains unchanged after 60 s of trypsin treatment. Scale bar: 10 μm. (E) GFP-Spire1C laterally diffuses along the mitochondrial outer membrane. A small region of a mitochondrion labeled with GFP-Spire1C was photobleached (white boxed region). The rapid, directional recovery from the unbleached region into the bleached region (see also Figure 2—figure supplement 2) suggests GFP-Spire1C is stably associated with and laterally diffuses along the mitochondrial outer membrane. (F) GFP-Spire1C does not readily exchange with the cytoplasm or neighboring mitochondria since photobleaching of an entire mitochondrion resulted in very low fluorescence recovery over the same period of time as in (E). Scale bar: 1 μm.
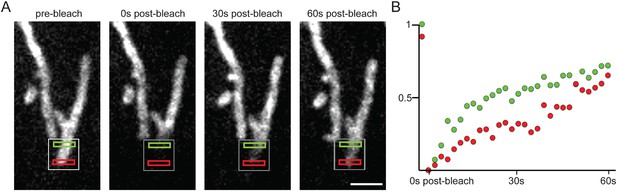
GFP-Spire1C laterally diffuses on the mitochondrial outer membrane.
(A) After partial bleaching of mitochondria labeled with GFP-Spire1C, rapid fluorescence recovery occurs in a directional fashion. The fluorescence intensity in the region closest to the unbleached region (green rectangle) increases more rapidly than the fluorescence intensity in the region farther away (red rectangle) from the unbleached region. Scale bar: 2 μm. (B) Graph of the fluorescence intensity of the green and red regions in (A) shows the differential rate of recovery over time for each region.
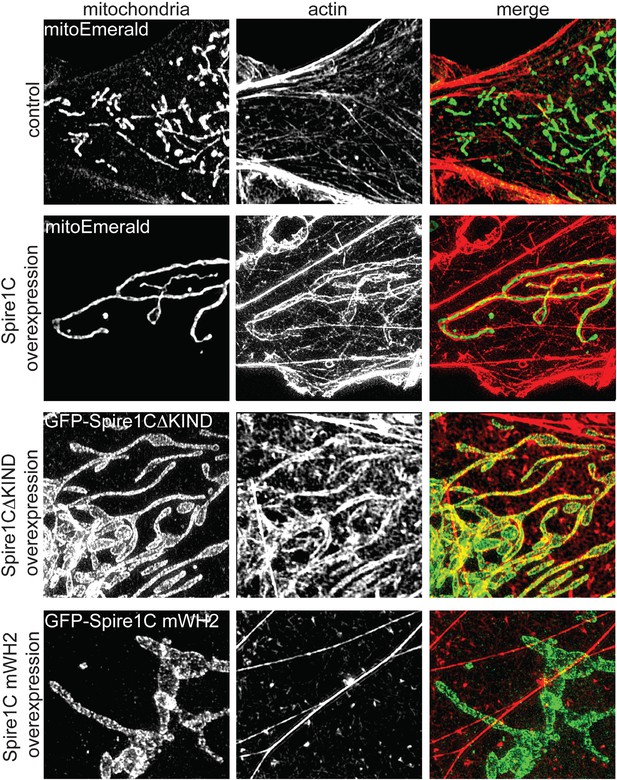
Spire1C promotes actin assembly on mitochondrial surfaces.
Overexpression of Spire1C causes actin accumulation on mitochondria. Control: SIM image of a Cos7 cell expressing mitoEmerald and stained with phalloidin-568 to visualize actin shows low amounts of overlap between mitochondria and actin (Mander's: 0.43 ± 0.020, ncells = 26). Spire1C overexpression: A Cos7 cell expressing mitoEmerald and overexpressing myc-Spire1C and stained with phalloidin-568 reveals significantly increased actin accumulation on mitochondria (Mander's: 0.64 ± 0.066, ncells = 19, p < 0.05) compared to control cells. GFP-Spire1CΔKIND: A Cos7 cell overexpressing the formin-binding deficient GFP-Spire1CΔKIND stained with phalloidin-568 reveals significant accumulation of actin on mitochondria compared to control cells (Mander's: 0.55 ± 0.017, ncells = 18, p < 0.05). A Cos7 cell overexpressing GFP-Spire1C mWH2 displays no increased accumulation of actin (Mander's: 0.41 ± 0.040, ncells = 15, p = 0.32) compared to control cells. Scale bar: 5 μm.
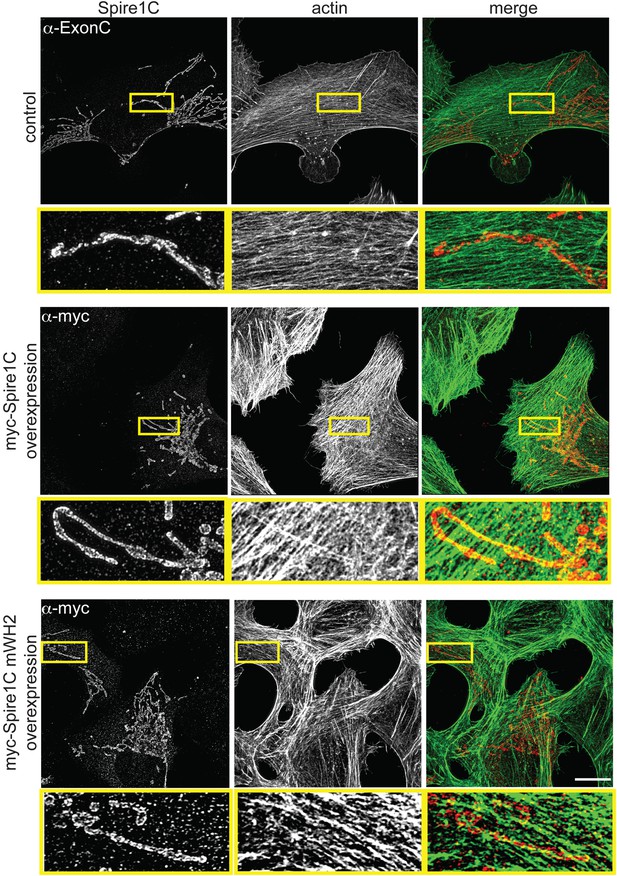
Spire1C promotes actin assembly near mitochondria in a WH2-dependent fashion.
control (α-ExonC): SIM image of a U2OS cell stained with phalloidin-488 and α-Spire1C to visualize endogenous Spire1C localization relative to actin shows very low amounts of overlap between mitochondria and actin. myc-Spire1C (α-myc): A U2OS cell overexpressing myc-Spire1C, stained with anti-myc and phalloidin-488 reveals increased actin accumulation on mitochondria compared to non-transfected cells. myc-Spire1C mWH2 (α-myc): A U2OS cell overexpressing the actin-nucleation deficient myc-Spire1C mWH2 labeled with anti-myc and phalloidin-488 reveals muted accumulation of actin on mitochondria compared to wild-type myc-Spire1C overexpression. Scale bars: 15 μm. The yellow-boxed regions are magnified underneath each image. All cells were counterstained with Alexa-568 secondary antibody.
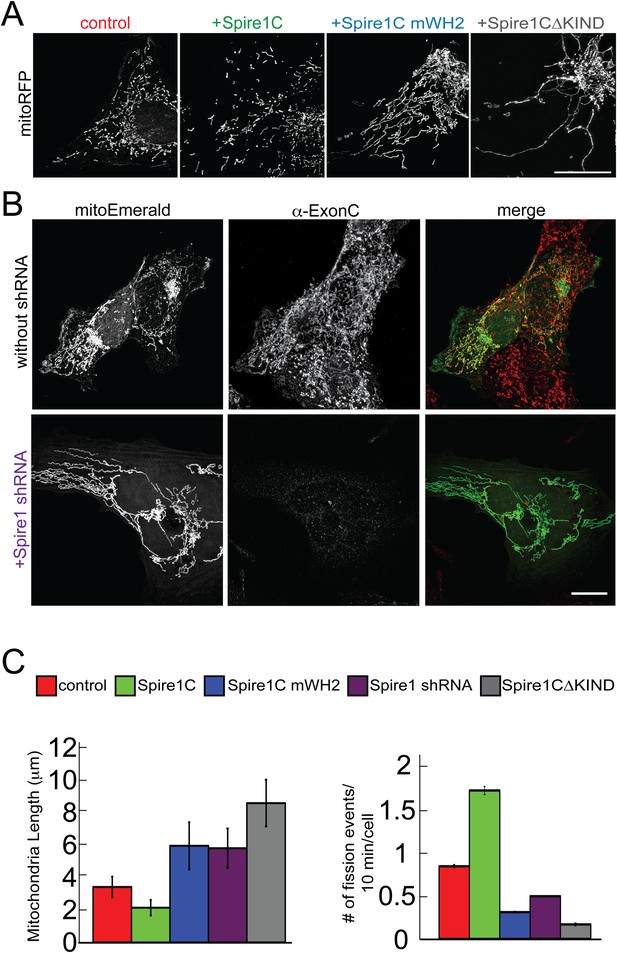
Spire1C promotes mitochondrial fission via its formin-binding KIND and actin-nucleating WH2 domains.
(A) U2OS cells overexpressing Spire1C display shorter mitochondria (second panel, 2.2 ± 0.5 μm, nmitochondria = 211, ncells = 14, p < 0.0001), whereas cells overexpressing Spire1C mWH2 (third panel, 6.2 ± 1.52 μm, nmitochondria = 332, ncells = 15, p < 0.0001) or Spire1CΔKIND (fourth panel, 9.0 ± 1.50 μm, nmitochondria = 232, ncells = 16, p < 0.0001) display longer, more tubulated mitochondria compared to control cells (first panel, 3.57 ± 0.45 μm, nmitochondria = 322, ncells = 34). Scale bar: 10 μm. (B) Cells transfected with mitoEmerald (and neighboring non-transfected cells) stained with α-ExonC (upper row) showed robust colocalization of mitoEmerald and α-ExonC, with a mixture of tubulated and fragmented mitochondria. Cells cotransfected with Spire1 shRNA and mitoEmerald with no detectable α-ExonC labeling (lower row) display long, tubulated mitochondria (6.1 ± 1.26 μm, nmitochondria = 222, ncells = 17). All primary antibodies were counterstained with Alexa-568 secondary antibody. Scale bar: 15 μm. (C) Left: Average mitochondrial lengths for control cells and cells overexpressing Spire1C, Spire1C mWH2, Spire1 shRNA or Spire1CΔKIND. Right: Average number of mitochondrial fission events in one cell in a timespan of 10 min for control (ncells = 17), Spire1C overexpressing (ncells = 10, p < 0.0001), Spire1C mWH2 overexpressing (ncells = 12, p < 0.0001), Spire1 knockdown (ncells = 25, p < 0.0001) and Spire1CΔKIND overexpressing (ncells = 22, p < 0.0001) cells. At least 3 separate experiments were performed for all conditions. Error bars represent standard error of the mean.
Distribution of mitochondrial lengths measured in each condition.
Histogram showing distribution of frequencies of mitochondrial lengths for each of the conditions shown in the bar graphs in Figure 4.
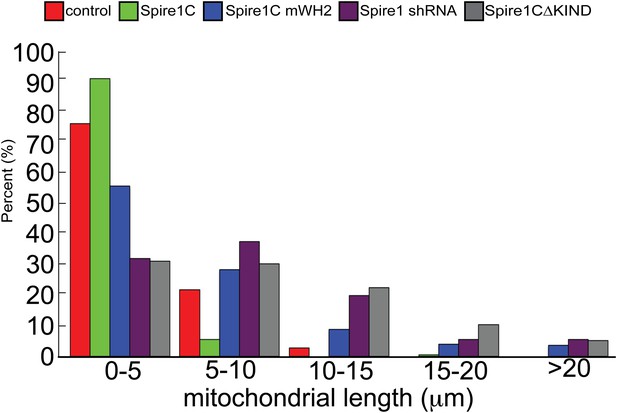
Spire1CΔKIND overexpression reduces the amount of ER-mitochondria overlap.
(A) Confocal images of cells expressing Ii33-mCherry and overexpressing GFP-Spire1C (second row) or GFP-Spire1C mWH2 (third row), but not GFP-Spire1CΔKIND (fourth row), display significant overlap of mitochondria with ER, similar to control cells expressing mitoEmerald. The images on the right-hand side show a magnified view of the boxed region in the merge image, with overlapping pixels in displayed in white. Scale bar: 15 μm. (B) Bar graph representing the average number of ER-mitochondria intersections per cell. We were able to resolve an average of 14.3 ± 3.5 intersections in control cells (ncells = 12). GFP-Spire1C overexpressing cells had 14.7 ± 1.93 (ncells = 15) ER-mitochondria intersections per cell. GFP-Spire1C mWH2 expressing cells had an average of 14.7 ± 1.62 (ncells = 11) ER-mitochondria intersections per cell. Spire1CΔKIND expressing cells had 6.8 ± 1.33 (ncells = 14, p < 0.05) ER-mitochondria intersections per cell.
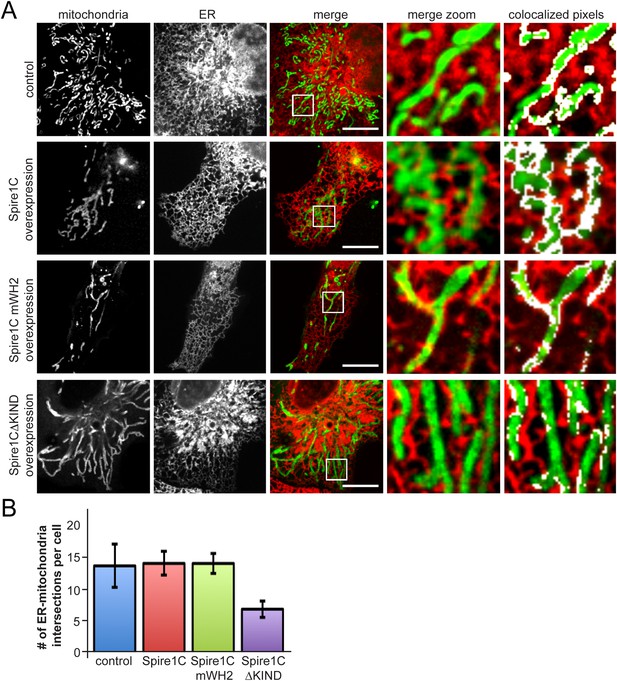
Spire1C and inverted formin 2 (INF2) directly interact and work together to regulate mitochondrial fission.
(A) INF2-CT directly binds to Spire1-KIND in vitro. GST pull-down assays in actin polymerization buffer, containing combinations of the following: 20 μM GST or GST-Spire1 KIND bound to glutathione-sepharose beads; 1 μM INF2-CT; and 10 μM INF2-NT. Co-incubation of the GST-Spire1 KIND domain pulls down INF2-CT (third to last lane), but not INF2-NT (second to last lane). INF2-CT pulldown is inhibited by the addition of the INF2-NT (last lane). STDS lane represents 0.2 μM INF2-CT. (B) Fluorescence anisotropy binding curve of purified INF2-CT (20 nM) labeled with tetramethylrhodamine succinimide mixed with varying concentrations of Spire1 KIND or bovine serum albumin reveals a direct interaction between Spire1 KIND and INF2-CT. (C) Cells overexpressing a constitutively active INF2 mutant (INF2 A149 alone, ncells = 16, nmitochondria = 232) display very short, fragmented mitochondria compared to control cells (p < 0.0001). Cells overexpressing A149 and Spire1C (+Spire1C, ncells = 14, nmitochondria = 461) or Spire1C mWH2 (+Spire1C mWH2, ncells = 20, nmitochondria = 377) similarly display very short mitochondria. In contrast, cells overexpressing A149 and Spire1CΔKIND display longer, more tubulated mitochondria (+Spire1CΔKIND, ncells = 18, nmitochondria = 379, p < 0.0001). (D) Cells overexpressing Spire1C and treated with scrambled siRNA display shorter, more fragmented mitochondria (+scramble siRNA, ncells = 20, nmitochondria = 434, p < 0.05). In contrast, cells overexpressing Spire1C and treated with INF2 siRNA display significantly longer, more tubulated mitochondria (Spire1C + INF2 siRNA, ncells = 22, nmitochondria = 627, p < 0.0001). Scale bars: 5 μm. (E) Bar graph displaying average mitochondria lengths for each of the conditions in this figure. Error bars represent standard error of the mean.
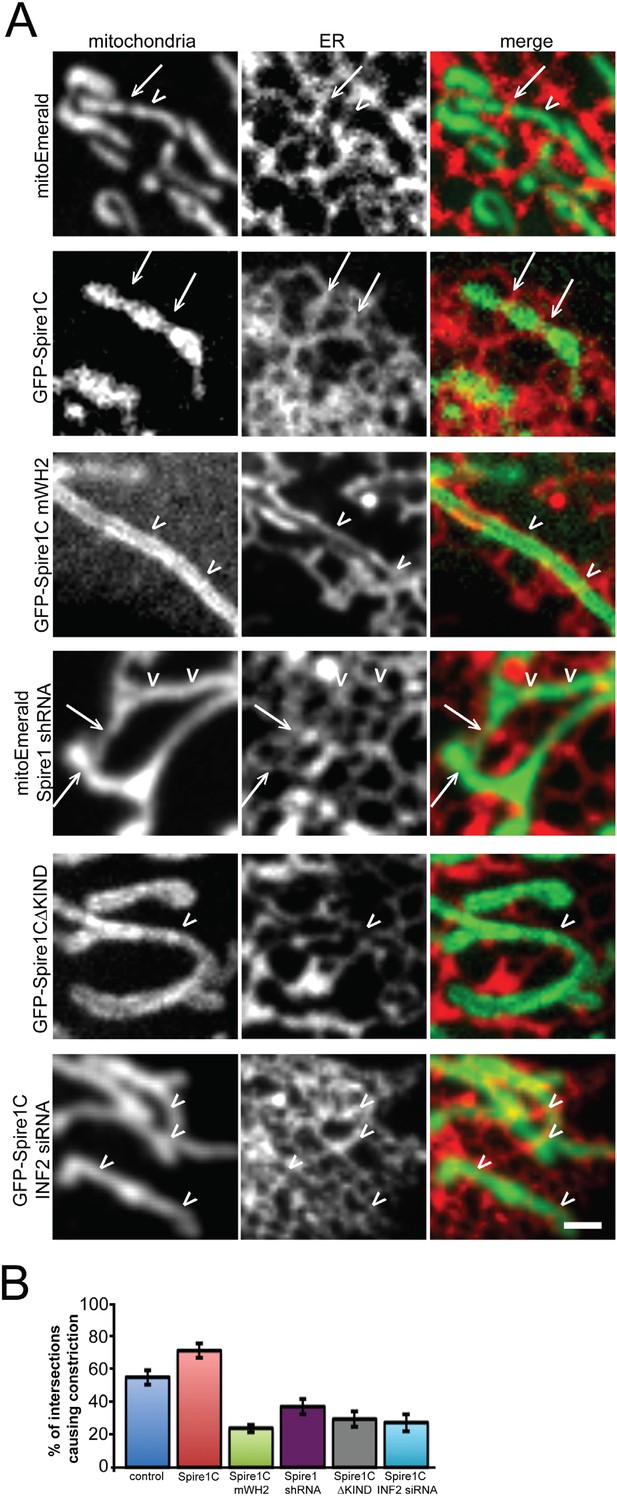
Spire1C overexpression enhances mitochondrial constriction via its WH2 and KIND domains in cooperation with INF2.
(A) Representative confocal images of U2OS cells expressing Ii33-mCherry in order to visualize ER tubules crossing over mitochondria in cells expressing mitoEmerald (first row) or overexpressing GFP-Spire1C (second row), GFP-Spire1C mWH2 (third row), GFP-Spire1CΔKIND (fourth row), or GFP-Spire1C while treated with INF2 siRNA (fifth row). Arrows indicate ER-mitochondria intersection points associated with mitochondrial constriction. Arrowheads indicate ER-mitochondria intersections not resulting in mitochondrial constriction. Scale bar: 1 μm. (B) Bar graphs representing the average percentage of ER-mitochondria intersections associated with mitochondrial constriction for each construct used. In cells expressing mitoEmerald, 55.2 ± 5.5% (nintersections = 172, ncells = 12) of ER-mitochondria intersections appeared to result in mitochondrial constriction. In GFP-Spire1C overexpressing cells, 71.5 ± 4.5% (nintersections = 221, ncells = 15, p < 0.05) of ER-mitochondria intersections resulted in mitochondrial constriction. In GFP-Spire1C mWH2 overexpressing cells, 24.1 ± 2.4% (nintersections = 162, ncells = 11, p < 0.01) of ER-mitochondria intersections appeared to result in mitochondrial constriction. In Spire1C knockdown cells, 37.2 ± 5.7% (nintersections = 123, ncells = 11, p < 0.001) of ER-mitochondria intersections appeared to result in mitochondrial constriction. In GFP-Spire1CΔKIND overexpressing cells, 29.5 ± 4.7% (nintersections = 95, ncells = 14, p < 0.01) of ER-mitochondria intersections appeared to result in mitochondrial constriction. In GFP-Spire1C overexpressing cells treated with INF2 siRNA, 27.5 ± 5.3% (nintersections = 178, ncells = 16, p < 0.01) of ER-mitochondria intersections appeared to result in mitochondrial constriction.
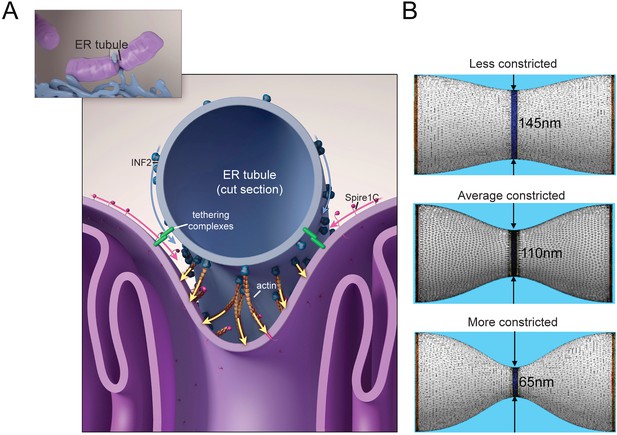
Putative model for how mitochondrial Spire1C and ER-anchored INF2 could mediate mitochondrial constriction via actin filament assembly.
(A) Spire1C:actin complexes on mitochondria associate with INF2 on the ER. Actin filaments nucleated by Spire1C are elongated by the actin polymerization activity of INF2. The actin filament elongation activity exerts pressure on the mitochondrial outer membrane, thereby driving constriction of the latter. Tethering complexes may play a role in maintaining association between ER and mitochondrial membranes. Myosin-II dimers and the related contractile actin ring, which may also be involved in mitochondrial constriction, are not shown for simplicity. (B) Computational results showing mitochondrial shapes resulting from deformation by constricting pressure P developed by the actin polymerization and/or actin contractile based mechanisms (see also Figure 8—figure supplement 1, Figure 8—source data 1, and ‘Materials and methods’ for more information). The mitochondrial constriction site was modeled as a tubular membrane of about 680 nm length and with initial radius R = 230 nm. The dark blue strip in the middle represents the 50 nm wide zone of the pressure application. The images correspond to 3° of the mitochondria constriction characterized by cross-sectional radii r in the narrowest place of 145 nm, 110 nm and 65 nm. The corresponding values of the pressure P, the required numbers of the polymerizing actin filaments, Nf, and the required tensions in the actin contractile ring, γm, are presented in Figure 8—figure supplement 1 and Figure 8—source data 1.
-
Figure 8—source data 1
Specific values of the system parameters and the computational results for the three specific extents of mitochondrial constriction presented in Figure 8, Figure 8—figure supplement 1, and discussed in the main text.
- https://doi.org/10.7554/eLife.08828.020
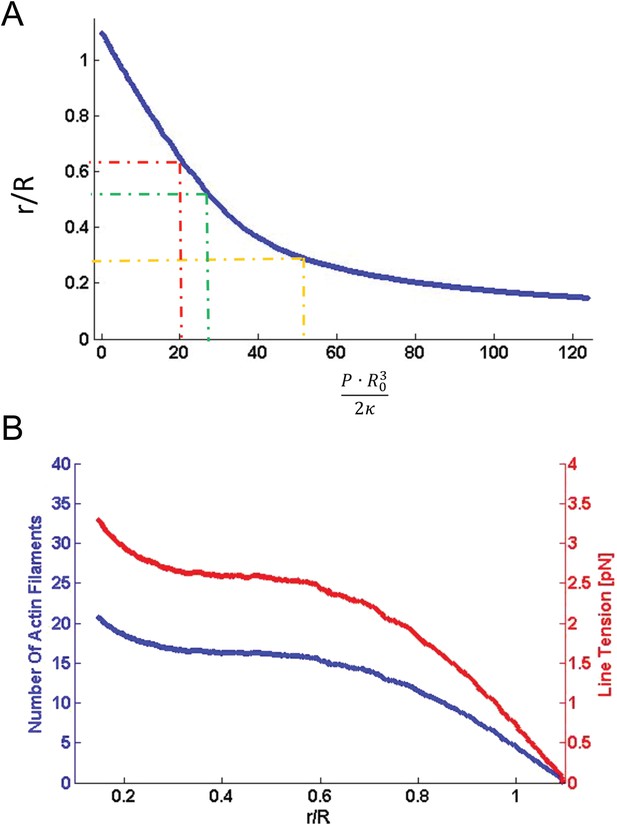
Computational results of simulations of mitochondrial constriction mediated by actin polymerization and actin constriction mechanisms.
(A) The cross-sectional radius in the narrowest place of the mitochondria shape, r, as a function of the pressure, P, exerted on the limited region in the middle of the constriction region (see Figure 8 of the main text). The radius, r, and the pressure, P, are presented in the universal dimensionless forms, r/R, and PR3/2κ, where R is the initial (preceding the deformation) mitochondrial radius and κ is the membrane bending modulus. The dashed lines indicate the specific deformations presented in Figure 8 of the main text. (B) The number of the actin filaments, Nf, and the tension in the actin contractile ring, γm, providing the pressure as functions of the resulting mitochondria deformation. The deformation is quantified by r/R (see (A) for definition).
Videos
A U2OS cell coexpressing GFP-Spire1C (N-terminus tag) and OMI-mCherry displays rapid loss of GFP fluorescence signal after the addition of 10 μM digitonin and 4 mM trypsin, whereas mCherry fluorescence persists, indicating that trypsin is degrading the GFP tag on the N-terminus of Spire1C in the cytoplasm, but not OMI-mCherry, which resides in the mitochondria intermembrane space (IMS).
Scale bar: 10 μm.
A U2OS cell coexpressing GFP-ExonC (N-terminus tag) and OMI-mCherry displays rapid loss of GFP fluorescence signal after the addition of 10 μM digitonin and 4 mM trypsin, whereas mCherry fluorescence persists, indicating that trypsin is degrading the GFP tag on the N-terminus of Spire1C in the cytoplasm, but not OMI-mCherry, which resides in the mitochondria IMS.
Scale bar: 10 μm.
A U2OS cell coexpressing ExonC-GFP (C-terminus tag) and OMI-mCherry displays no loss of GFP fluorescence signal after the addition of 10 μM digitonin and 4 mM trypsin, indicating the GFP tag on the C-terminus of ExonC is protected within the mitochondrial lumen.
Scale bar: 10 μm.





