Physiological modulation of BiP activity by trans-protomer engagement of the interdomain linker
Figures
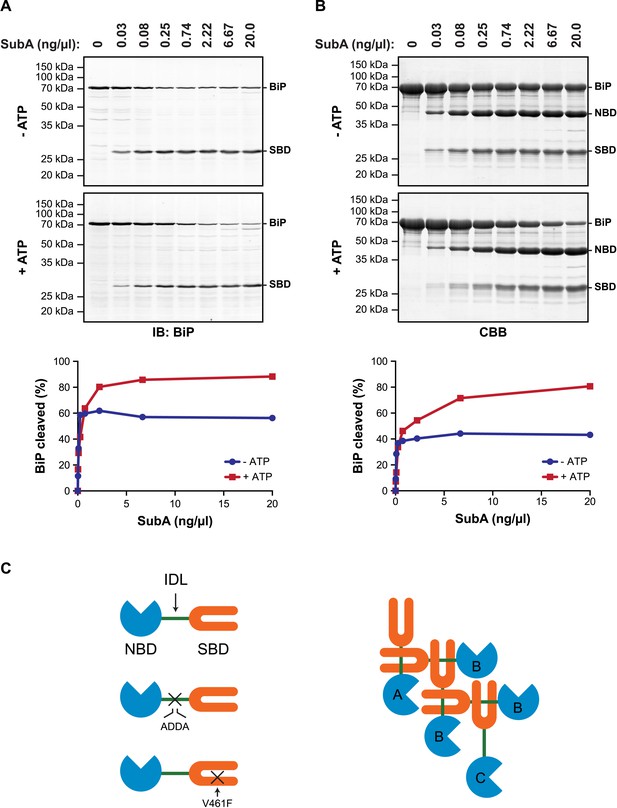
ATP increases sensitivity of BiP to cleavage at its interdomain linker.
(A) Immunoblot of endogenous BiP in lysates of CHO-K1 cells, following exposure (10 min at 30°C) to the indicated concentration of a bacterial protease (SubA) that selectively cleaves the interdomain linker and separation by SDS-PAGE. The lysate in the upper panel was prepared in presence of hexokinase and glucose to deplete ATP. The lysate in the lower panel was prepared without hexokinase and exposed to ATP (1.5 mM) before cleavage by SubA. The full-length protein (BiP) and the C-terminal substrate binding domain (SBD) were detected with antiserum that recognizes a C-terminal epitope of hamster BiP, quantified by fluorescence-based immunoblotting on a Licor Odyssey imager and displayed in graphic form in the bottom panel. (B) Purified BiP (12 µM) was incubated (10 min at 30°C) with or without 3 mM ATP followed by exposure (20 min at 30°C) to the indicated concentration of SubA. The samples were then analyzed by SDS-PAGE and Coomassie (CBB)-staining. The intact protein (BiP), nucleotide binding domain (NBD) and SBD are indicated. Signal intensity was quantified by detecting Coomassie fluorescence on a Licor Odyssey imager and displayed in graphic form in the bottom panel. Shown are representative experimental observations, reproduced three times. (C) Cartoon describing the proposed architecture of BiP oligomers: The SBD is in orange, the NBD in blue and the interdomain linker (IDL) in green. The sites of the ADDA mutation that abolishes SubA cleavage and alters the character of the interdomain linker and that of the V461F mutation that interferes with substrate binding are indicated (crosses). The oligomer is predicted to encompass protomers with three different dispositions. Protomer A is allosterically uncoupled but possesses a free SBD capable of engaging substrates (or other BiP molecules through their interdomain linker). Protomer B is allosterically uncoupled and unable to bind substrates. Protomer C is unable to bind substrates but is subject to allosteric regulation via its NBD.
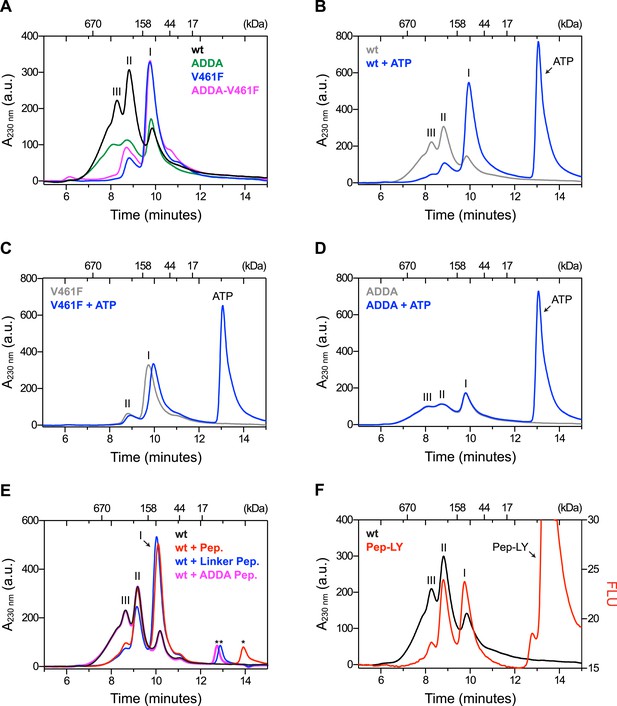
Mutations that disrupt substrate binding or alter the interdomain linker interfere with BiP oligomerization.
(A) Peptide bond absorbance traces (A230 nm) of solutions of bacterially expressed and purified wildtype (wt) BiP, or the indicated mutants (all at 50 µM), following fractionation by size-exclusion chromatography (SEC-3 column). The distinct peaks eluting at 9.85 min (I), 8.82 min (II) and 8.28 min (III) are marked. The ADDA mutation (L414VLL417 to A414DDA417) inactivates the linker's role in interdomain allostery (Swain et al., 2007; Kumar et al., 2011), whereas the V461F mutation inhibits substrate binding (Petrova et al., 2008). Note relatively fewer oligomeric species (fractions III and II) in the preparations of mutant proteins. (B) Trace as in ‘A’ of wildtype BiP incubated in the absence or presence of 5 mM ATP before fractionation. The BiP profile described in ‘A’ and the profile of BiP incubated with ATP are shown. The additional peak at 13.07 min represents free ATP. Note the dissociation of oligomers (from peaks III and II to peak I) upon exposure to ATP. (C) Trace as in ‘B’ with the V461F mutant BiP. (D) Trace as in ‘B’ with the ADDA mutant BiP. (E) Wildtype BiP (50 µM) was incubated without or with a BiP-binding peptide (HTFPAVL, ‘Pep.’; 1 mM) (Marcinowski et al., 2011) or a peptide derived from the interdomain linker of BiP (DTGDLVLLD, ‘Linker Pep.’; 1 mM) or a mutant linker peptide (DTGDADDAD, ‘ADDA Pep.’; 1 mM) and fractionated as in ‘A’. Note that whereas addition of both the reference peptide (HTFPAVL) and the linker peptide (DTGDLVLLD) shifted the distribution of BiP to lower molecular weight species, the mutant linker peptide (DTGDADDAD) was with no effect. The asterisks mark the elution peaks of free peptides. (F) Wildtype BiP (50 µM) was incubated in the absence or presence of tracer amounts lucifer yellow-labeled BiP-binding peptide (HTFPAVL-LY, 'Pep-LY'; 1 µM) and fractionated as in ‘A’. Absorbance (A230 nm; black) and the fluorescence trace (in arbitrary fluorescence units, FLU) of lucifer yellow (LY) (Ex: 430 nm, Em: 525 nm; red) are shown.
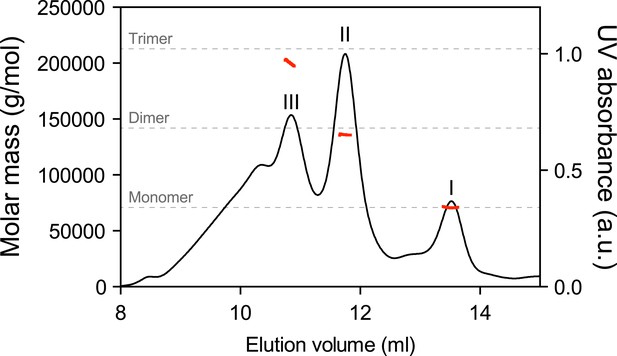
Multiangle light scattering (SEC-MALS) measurement of purified BiP eluted from a size-exclusion chromatography column.
Purified wildtype BiP (90 µM) was passed through a size-exclusion chromatography (SEC) column and the molar masses of BiP complexes in elution peaks I, II and III were measured by MALS. Weight-averaged molar masses (solid red lines) are shown across the elution profile (UV absorbance at 280 nm, black line). The predicted masses of BiP monomers, dimers and trimers are indicated (grey dashed lines).
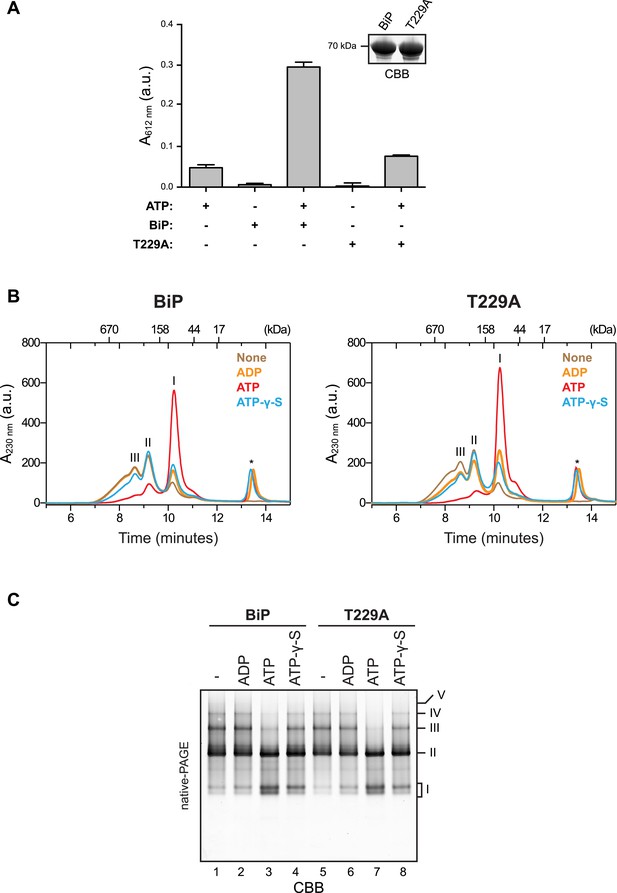
ATP binding rather than ATP hydrolysis induces dissociation of BiP oligomers.
(A) Colorimetric measurement of ATP hydrolysis by purified wildtype BiP and the ATPase activity-deficient mutant BiPT229A. Shown are mean ± SD of three technical replicates (n = 3). Samples of the BiP protein working solutions were applied to SDS-PAGE and visualized by Coomassie staining (inset) to confirm that equal protein amounts were used in the assay. (B) Size-exclusion chromatography (SEC) analysis of wildtype BiP and BiPT229A. BiP proteins were diluted to 50 µM in absence or presence of 800 µM ATP, ADP or ATP-γ-S, respectively, and incubated for 20 min at room temperature before injection onto a SEC column. The asterisks mark the elution peaks of free nucleotides. Note that ATP induced dissociation of oligomeric BiP species. ADP caused partial decrease of oligomers formed by BiPT229A but had almost no effect on wildtype BiP oligomers. ATP-γ-S caused only partial disassembly of wildtype and mutant BiPT229A oligomers, which is consistent with the previously-observed inefficient dissociation of Hsp70:substrate complexes by this ATP analog (Palleros et al., 1993). (C) Effect of nucleotides on BiP oligomers analyzed by native-PAGE. BiP or BiPT229A were incubated with the indicated nucleotides prior to separation on a native gel and Coomassie staining. The major species are numbered by order of descending mobility (I-V). Note the conversion of slower migrating species (III-V) into faster migrating ones (I and II) by incubation with ATP and to a lesser extend with ATP-γ-S. Also note that the nucleotide-dependent differences in the oligomerization status of BiP appear weaker on native gels compared to SEC analysis in ‘B’, which is likely due to re-formation of oligomeric complexes caused by nucleotide depletion and protein concentration effects during native-PAGE.
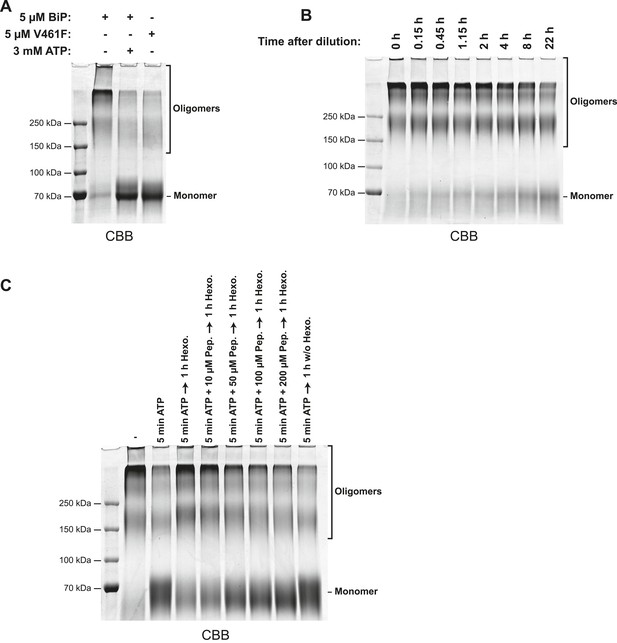
Glutaraldehyde crosslinking confirms the nucleotide dependence of BiP oligomerization and its susceptibility to competition by substrate peptides.
(A) SDS-PAGE and Coomassie staining of purified wildtype BiP or substrate binding-deficient BiPV461F following brief crosslinking with gluteraldehyde. Protein samples (at 10 µM) were pre-incubated for 16 hr at 25°C in HKM buffer and challenged with ATP (3 mM for 10 min; where indicated) before dilution (to 0.18 µM final protein concentration) in HKM containing glutaraldehyde (0.05% final) and further incubated for 2 min at 25°C. The crosslinking reactions were quenched on ice with 80 mM sodium borohydride and 200 mM Tris and proteins were TCA precipitated and analyzed by SDS-PAGE (7% gels) and Coomassie staining. Monomeric BiP and slower-migrating oligomeric crosslinked complexes are indicated. Note that wildtype BiP oligomers disassembled into monomers in presence of ATP, whereas BiPV461F remained largely monomeric even in absence of added nucleotide. (B) Coomassie-stained SDS-gel of gluteraldehyde crosslinked wildtype BiP following equilibration at high concentration (50 µM), dilution to a final concentration of 0.18 µM in HKM buffer and to re-equilibration at this low concentration for the indicated times before glutaraldehyde crosslinking, quenching, concentration by TCA precipitation and analysis as described in ‘A’. Note the slow dissociation of BiP oligomers into monomers upon dilution. (C) Coomassie-stained SDS-gel of gluteraldehyde crosslinked wildtype BiP as above. Purified wildtype BiP (10 µM) was incubated without or with 2 mM ATP and BiP-binding peptide (HTFPAVL, 'Pep.') at the indicated concentrations in HKM buffer containing 15 mM D-glucose for 5 min at 30°C to allow ATP-mediated dissociation of BiP oligomers. ATP was then hydrolyzed with 0.3 mU/µl hexokinase for 1 hr at 30°C to allow re-formation of BiP oligomers in the absence or presence of competing substrate peptide followed by glutaraldehyde crosslinking as in ‘A’. Note that substrate peptides interfered with the formation of BiP oligomers (during ATP-depletion) in a concentration-dependent manner.
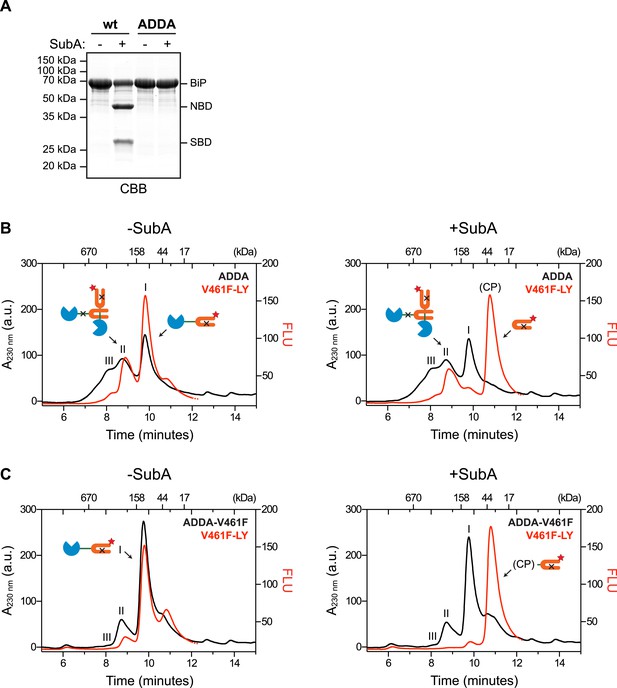
Cross-protomer engagement of the interdomain linker in BiP oligomers protects it against cleavage by SubA.
(A) Coomassie-stained SDS-gel of wildtype (wt) and ADDA mutant BiP (12 µM) before and after cleavage with SubA (0.25 ng/µl) for 15 min. Note that the ADDA mutation precludes cleavage by SubA. (B) Peptide bond absorbance trace (A230 nm, black) and fluorescence trace (in arbitrary fluorescence units, FLU) of lucifer yellow (LY) (Ex: 430 nm, Em: 525 nm; red) of purified ADDA mutant BiP (50 µM) incubated with tracer concentrations (1 µM) of lucifer yellow-labeled V461F mutant BiP (V461F-LY) and fractionated by size-exclusion chromatography (as in Figure 2) either before (-SubA) or after (+SubA) cleavage with SubA (36 ng/µl). Peaks I, II and III are marked as is the new fluorescent peak emanating from the V461F-LY cleavage product (CP). Note that fluorescent peak I, corresponding to the V461F mutant BiP monomer is attenuated after cleavage with SubA, whereas peak II, arising from hetero-oligomers of ADDA mutant BiP and V461F mutant BiP is relatively resistant. The peptide-bond absorbance trace (A230 nm), contributed largely by the ADDA mutant BiP, is unchanged by cleavage with SubA. (C) Experiment as in ‘B’ with purified double mutant ADDA-V461F BiP (50 µM) and lucifer yellow-labeled V461F mutant BiP (V461F-LY). Note the absence of a SubA-resistant fluorescent peak II in this sample.
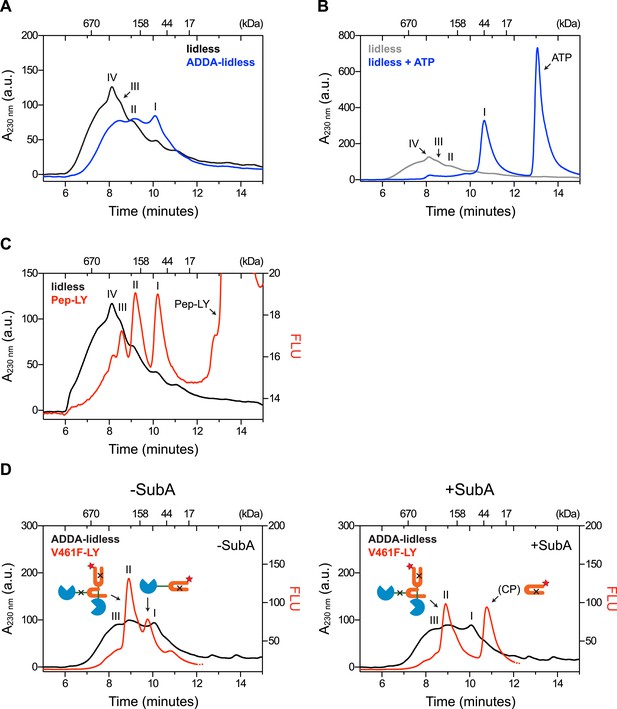
Removal of BiP's lid does not abrogate cross-protomer engagement of the interdomain linker in BiP oligomers.
(A) Peptide bond absorbance traces (A230 nm) of purified BiP (50 µM) lacking its C-terminal ‘lid’ (∆554–654) without (lidless) or with a compounding ADDA mutation (ADDA-lidless) following fractionation by size-exclusion chromatography (SEC-3 column). The distinct peaks I, II and III (described in the legend to Figure 2) are indicated. (B) Trace as in ‘A’ of lidless BiP incubated with or without ATP (5 mM) before fractionation. Note the dissociation of oligomers upon exposure to ATP. (C) Peptide bond absorbance trace (A230 nm, black) and lucifer yellow (LY) fluorescence trace (Ex: 430 nm, Em: 525 nm; red) of purified lidless BiP (50 µM; as in ‘A’) supplemented with tracer concentrations of LY-labeled BiP-binding peptide (1 µM; as in Figure 2E) and fractionated as in ‘A’. The peak corresponding to the free peptide is indicated. Note the disproportionately diminished engagement of peptide by the larger lidless BiP oligomers, (D) Peptide bond absorbance trace of purified ADDA-lidless mutant BiP (50 µM) (A230 nm, black) and fluorescence trace (red) of tracer concentrations (1 µM) of fluorescent LY-labeled V461F mutant BiP (V461F-LY) fractionated by size-exclusion chromatography either before (-SubA) or after (+SubA) cleavage with SubA (36 ng/µl; as in Figure 3B). Peaks I, II and III are marked as is the new fluorescent peak emanating from the V461F-LY cleavage product (CP). Note that fluorescent peak I is attenuated upon treatment with SubA, whereas peak II is resistant. The peptide-bond absorbance trace, contributed largely by the ADDA mutant lidless BiP, is unchanged by SubA cleavage.
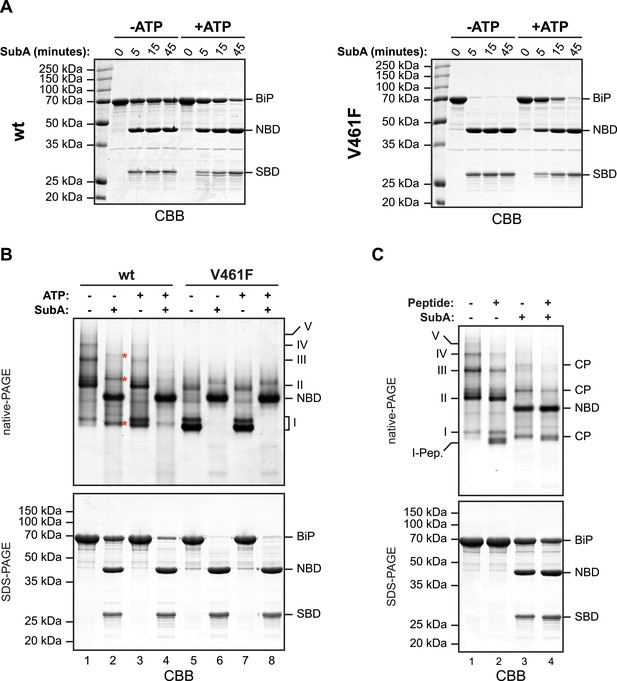
In oligomers, only a subset of protomers expose their interdomain linker to SubA.
(A) Purified wildtype (wt) and V461F mutant BiP (both at 7.5 µM) were exposed to SubA (20 ng/µl) at 30°C for the indicated time in presence or absence of 2 mM ATP and analyzed by SDS-PAGE and Coomassie staining. The intact protein (BiP), nucleotide binding domain (NBD) and substrate binding domain (SBD) are indicated. Note that ATP enhances cleavage of the wildtype BiP whilst attenuating cleavage of the V461F mutant. (B) Coomassie-stained native gel (upper panel) of purified wildtype and V461F mutant BiP (both at 50 µM). Where indicated the sample had been exposed to 5 mM ATP and SubA (15 ng/µl) and digested to completion. The major species have been numbered I-V by order of descending mobility. Red asterisks mark the new species emerging after digestion with SubA and the cleaved NBD is noted for reference. The same samples were applied to SDS-PAGE and the gel was stained with Coomassie (lower panel). (C) Wildtype BiP (50 µM) was incubated in presence or absence of 500 µM BiP-binding peptide (HTFPAVL [Marcinowski et al., 2011]) for 16 hr at room-temperature followed by digestion with SubA, as is ‘B’. The samples were then analyzed in parallel by native-PAGE (upper panel) and SDS-PAGE (lower panel) and the gels were stained with Coomassie. The novel species emerging after addition of peptide is labeled 'I-Pep' and the cleavage products of BiP (CP) are indicated. Note that both peptide addition and ATP shift the distribution of BiP oligomers to faster migrating species on native gels and enhance sensitivity to cleavage by SubA.
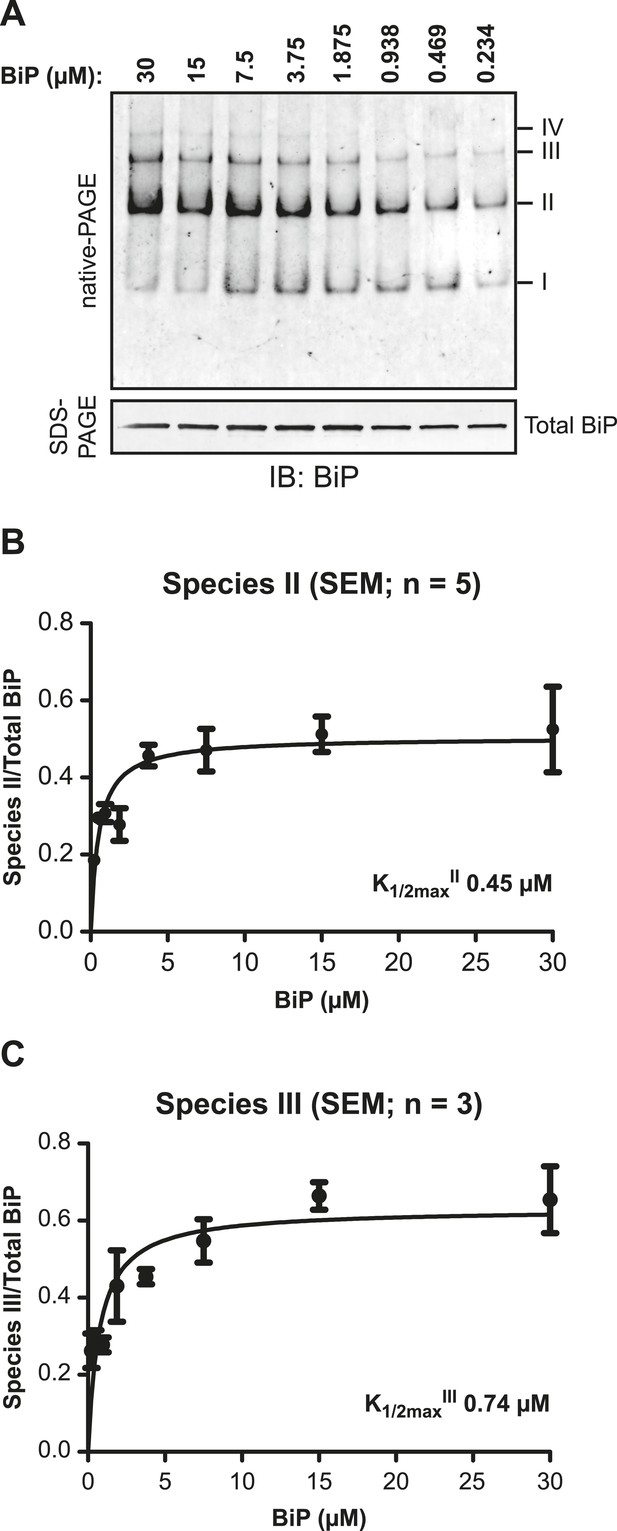
BiP protomers have high affinity for each other.
(A) Immunoblot of purified BiP resolved by native-PAGE. BiP was diluted to the indicated concentrations and allowed to equilibrate for 48 hr at room temperature. To obtain similar signal intensities in all lanes the samples were further diluted directly before gel electrophoresis and similar protein mass (0.25 µg) was loaded on each lane. The major species have been numbered I-IV by order of descending mobility. A parallel immunoblot of the same sample resolved by SDS-PAGE (lower panel) reports of the mass of BiP loaded. (B) Plot of the proportion of BiP in species II as a function of protein concentration. The signal of BiP species II was quantified and normalized to signal of total BiP loaded. (C) Plot of the proportion of BiP in species III as a function of protein concentration as in ‘B’.
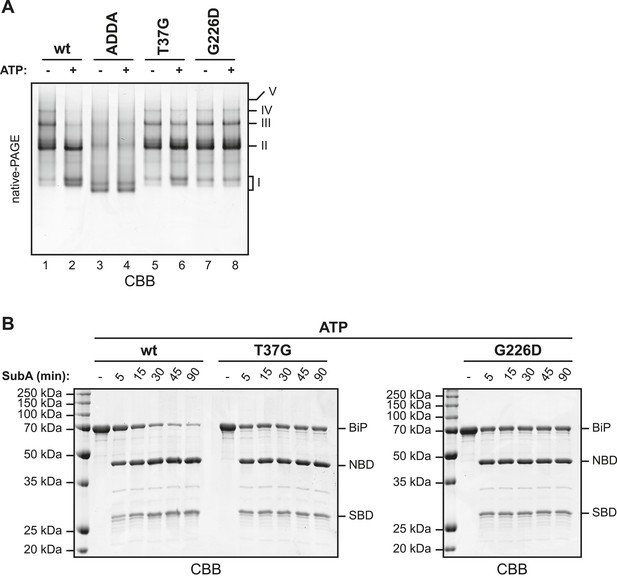
BiPADDA fails to form discrete oligomers in contrast to other BiP mutants that are locked in the domain-uncoupled state.
(A) Purified wildtype (wt) and mutant BiP proteins (BiPADDA, BiPT37G and BiPG226D) were incubated (all at 30 µM) for 15 min at 30°C with or without 3 mM ATP in HKM buffer and analyzed by native-PAGE and Coomassie-staining. The major species have been numbered I-V by order of descending mobility. Whereas BiPADDA is severely impaired in the formation of discrete oligomers, the other mutants, whose nucleotide binding domains (NBD) and substrate binding-domains (SBD) are locked in the uncoupled state, form oligomers that persist in presence of ATP. (B) Purified wildtype and mutant BiP proteins (all at 7.5 µM) were exposed to SubA (20 ng/µl) at 30°C for the indicated times in presence of 3 mM ATP and subsequently analyzed by SDS-PAGE and Coomassie staining. The full-length proteins (BiP) as well as NBD and SBD are indicated. Note that the mutant BiP proteins are less efficiently cleaved by SubA.
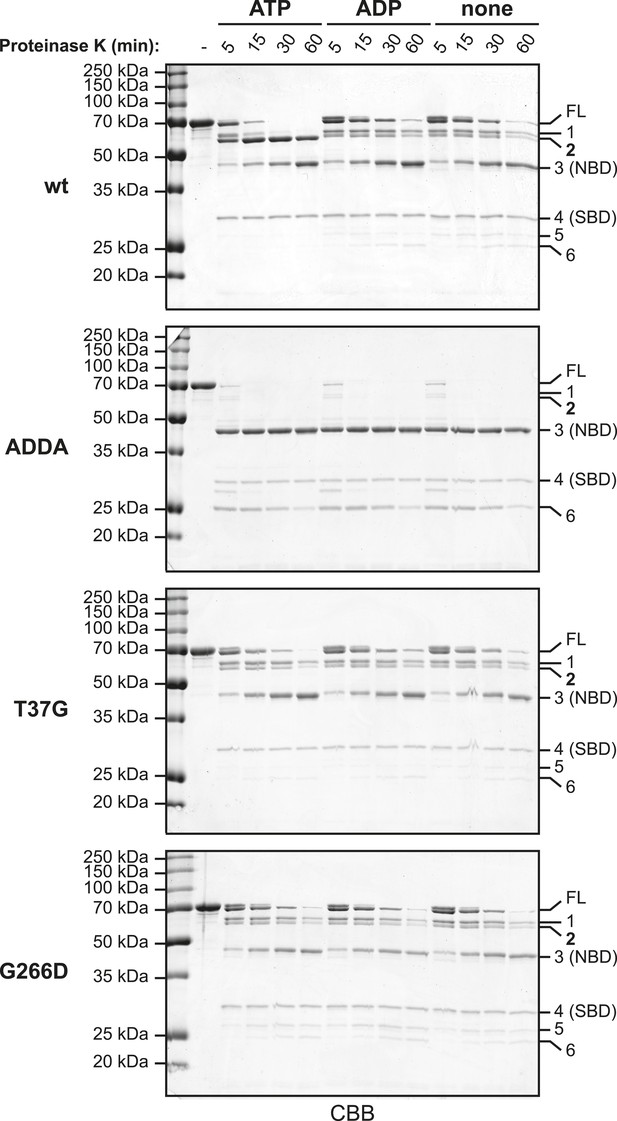
Partial proteolysis to analyze the nucleotide-dependent conformational states of wildtype and mutant BiP proteins.
10 µg of purified wildtype (wt) and mutant BiP proteins (BiPADDA, BiPT37G and BiPG226D) were digested with 2 µg Proteinase K in HKM buffer for the indicated times without or with added nucleotide (ATP or ADP; each 1.5 mM) and subsequently analyzed by SDS-PAGE and Coomassie-staining. Full-length proteins (FL) and cleavage fragments (1–6) are marked. Fragments 3 and 4 likely represent the compact nucleotide binding-domain (NBD; ∼ 44 kDa) and the substrate binding-domain (SBD; ∼ 27 kDa), respectively. Note the accumulation of a characteristic 60 kDa fragment (2) during digestion of wt BiP in presence of ATP, which reports on the ATP-induced compact conformation of the protein and protection of the interdomain linker. In contrast, neither of the mutants showed significant nucleotide-dependent changes of the proteolytic pattern. Also note the fast and complete digestion of full-length BiPADDA and the concomitant appearance of the 44 kDa fragment 3, suggesting that the linker was readily exposed to the protease and the NBD and SBD were uncoupled.
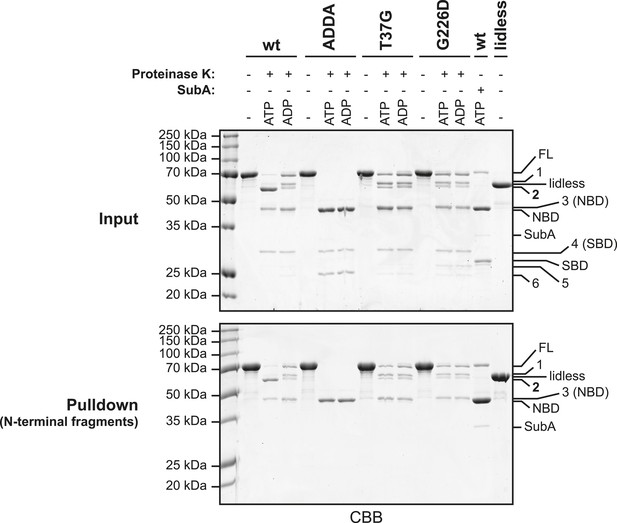
Characterization of the proteolytic fragments generated by digestion of BiP with Proteinase K.
Wildtype (wt) and mutant BiP proteins (BiPADDA, BiPT37G and BiPG226D) carrying N-terminal hexahistidine tags (His6-tag) were digested for 25 min with Proteinase K in presence or absence of added nucleotide (ATP or ADP; each 1.5 mM). Afterwards, samples were taken and proteins in the remaining reaction mixtures were denatured with 6 M guanidine hydrochloride (GuHCl) and bound to nickel-affinity beads (Ni-NTA agarose) in presence of 2 mM β-mecaptoethanol and 15 mM imidazole. The beads were washed with 4 M GuHCl in HKM buffer and the retained N-terminal BiP fragments were eluted with SDS sample buffer containing 250 mM imidazole. As controls wt BiP, digested with the linker-specific protease SubA, and undigested BiPlidless were affinity purified in parallel. The samples taken before affinity purification (Input) and the eluted polypeptides (Pulldown) were analyzed by SDS-PAGE and Coomassie staining. Full-length proteins (FL) and their proteolytic fragments (1–6) that resulted from digestion with Proteinase K are indicated. Note the similar migration of cleavage fragment 2 and BiPlidless, which lacks part of the C-terminal lid structure. In addition, the nucleotide binding domain (NBD) and the substrate binding domain (SBD), generated by digestion of wildtype BiP with SubA, migrate very similar to fragments 3 and 4, respectively. Also note that fragment 2 (which reports on the ATP-induced compact conformation of the protein) as well as fragments 1 and 3 represent N-terminal cleavage products, since they were efficiently retained on nickel-beads under denaturing conditions. This further implies that fragment 2 results from cleavage of BiP within the C-terminal lid of the open SBD, which is exposed in the ATP-bound conformation (Kityk et al., 2012; Qi et al., 2013).
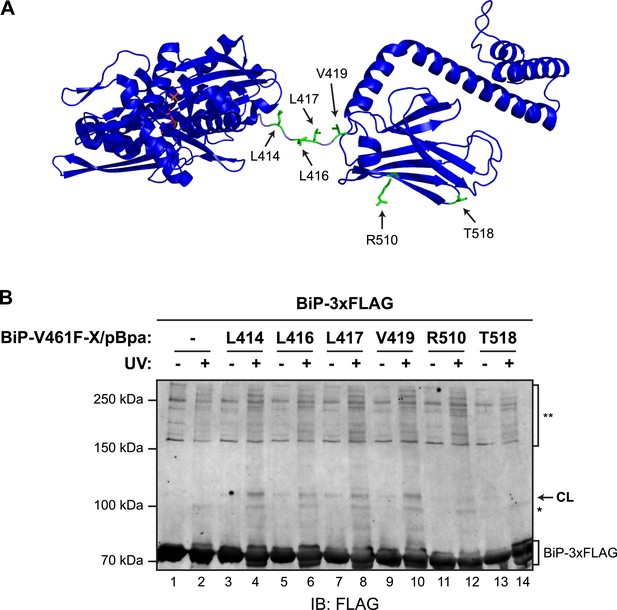
Site-specific photo-crosslinking to probe the trans-engagement of the interdomain linker in BiP oligomers.
(A) Residues of BiP that were individually replaced by the photoactivatable crosslinking amino acid p-benzoyl-L-phenylalanine (pBpa) are highlighted (green) on a model of BiP in the ADP-bound state that is based on the crystal structure of DnaK from Geobacillus kaustophilus (PDB: 2V7Y). (B) Anti-FLAG immunoblot of FLAG-tagged wildtype BiP (BiP-3xFLAG) from in vitro photo-crosslinking reactions resolved on a 7% SDS-gel. BiP-3xFLAG (0.2 µM final) was combined with untagged BiP-V461F-X/pBpa proteins (2 µM final) carrying pBpa at either one of the positions (X) illustrated in ‘A’. The mixtures were further incubated for 16 hr at 25°C to allow for efficient formation of trans BiP complexes. Where indicated samples were then exposed to UV light for 20 min on ice. The position of pBpa decorated BiP-dependent (‘specific’) crosslinked adduct is indicated (CL) as are the decorated BiP-independent (‘non-specific adducts’, *) and FLAG immunoreactive high-molecular weight material (**). Note that the specific UV-enhanced CL signals were diminished (or absent) when pBpa was incorporated at positions (aa 510 and 518) distant from the interdomain linker.
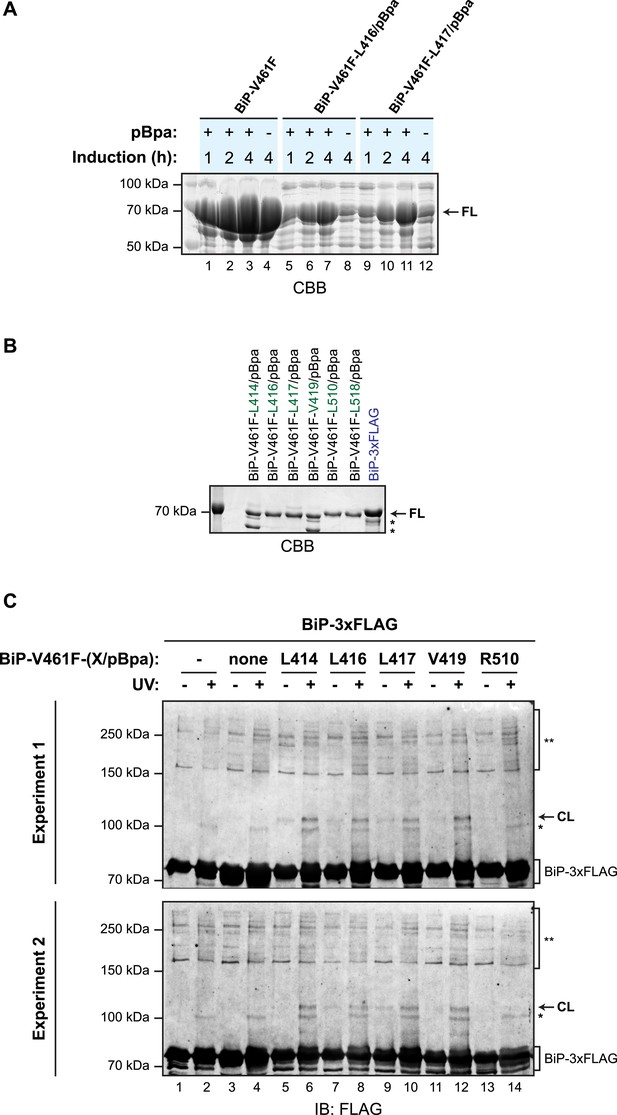
Site-specific photo-crosslinking yields a specific adduct.
(A) Coomassie-stained SDS-gel of otherwise identical C-terminally hexahistidine- (His6-) tagged BiPV461F expressed from plasmids lacking (lanes 1–4) or bearing site specific amber (TAG) codons replacing L416 (lanes 5–8) or L417 (lanes 9–12), partially-purified by nickel affinity chromatography and resolved on a 10% SDS-gel. Bacteria transformed with the aforementioned plasmids were induced for the indicated time with IPTG in the absence or presence of p-benzoyl-L-phenylalanine (pBpa). Note that the yield of BiP from bacteria transformed with plasmid lacking the amber codon was unaffected by the addition of pBpa to the culture, whereas the yield from cultures transformed with plasmids bearing amber codons was notably enhanced by addition of pBpa, as predicted (Ryu and Schultz, 2006). (B) Coomassie-stained SDS-gel of the BiP proteins used for site-specific crosslinking experiments shown in Figure 6B and below. (C) Immunoblots of FLAG-tagged wildtype BiP (BiP-3xFLAG) from independent repeats of the in vitro photo-crosslinking experiment as in Figure 6B. Lanes 1 and 2 contained only the 3xFLAG-tagged BiP and lanes 3 and 4 contained non-derivatized untagged BiP as controls.
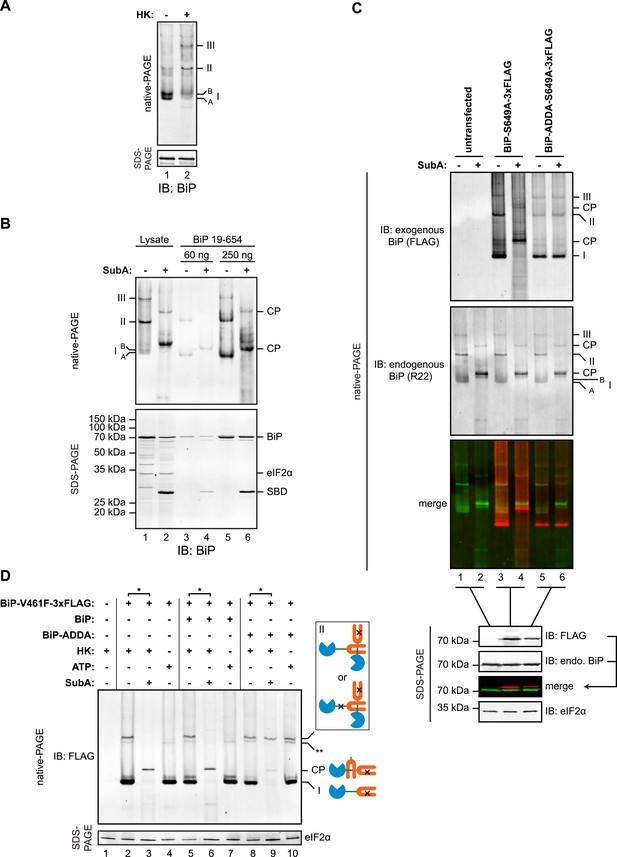
Trans-engagement of the linker by the substrate binding domain (SBD) as the basis for BiP oligomerization in vivo.
(A) Immunoblot of endogenous BiP from lysates of CHO-K1 cells resolved by native-PAGE. Where indicated the lysate was depleted of ATP by incubation with hexokinase and glucose (HK). The major species observed on the native gel have been numbered I-III by order of descending mobility and the two forms of BiP detected in the higher mobility region of the gel (‘A’ and ‘B’) are indicated. Immunoblot of the same sample resolved by SDS-PAGE (lower panel) serves as a loading control. (B) Immunoblot of endogenous BiP from CHO-K1 cells lysed in presence of hexokinase and glucose to deplete ATP (lysate) or mature hamster BiP (19–654) expressed in Escherichia coli and resolved on the same native gel. Where indicated the lysate and or pure BiP were exposed to SubA (5 ng/µl for 10 min at room temperature). The emergent cleavage products (CP) are indicated. Immunoblot of the same sample resolved by SDS-PAGE (lower panel). The free nucleotide binding domain (NBD) is undetected by this antiserum that recognizes the C-terminus of BiP. (C) Anti-FLAG and anti-BiP immunoblots of native gels of ATP-depleted lysates from CHO-K1 cells transfected with the indicated BiP constructs bearing an S649A mutation abolishing their reactivity with the anti-BiP R22 serum. The lysates were split and exposed to SubA as in ‘B’. Anti-FLAG, anti-endogenous BiP (R22) and anti-eIF2α (a loading control) immunoblots of the same samples resolved by SDS-PAGE are presented in the panels below. Note that the FLAG-tagged exogenous BiP proteins are not recognized by the polyclonal anti-rodent BiP antiserum due to the S649A substitution. (D) Anti-FLAG immunoblot of ATP-depleted lysates from CHO-K1 cells transfected with the indicated BiP derivatives and resolved by native-PAGE. Where indicated, the two samples from the same lysate (*) were treated with or without SubA as in ‘B’. ATP (3 mM) was added to lysates loaded in lanes 4, 7 and 10 (prepared without hexokinase and glucose) to dissociate oligomeric species. Anti-eIF2α immunoblot of the same samples resolved by SDS-PAGE is presented in the panel below as a loading control. To aid interpretation, a schema of the BiP species involved is provided to the side of the gel. The BiP NBD is colored blue its SBD orange and the interdomain linker green. The V461F mutation (in the SBD) and the ADDA mutation (in the interdomain linker) are indicated by crosses. Note that the BiP complexes represented by species II in lanes 9 and 10 are resistant to both, SubA and ATP. The identity of the band marked with ** is unknown. Note: To promote BiP oligomerization in vivo, the CHO-K1 cells in panels B-D were treated with thapsigargin (0.5 µM for 10 min before harvest, as will be explained in detail below).
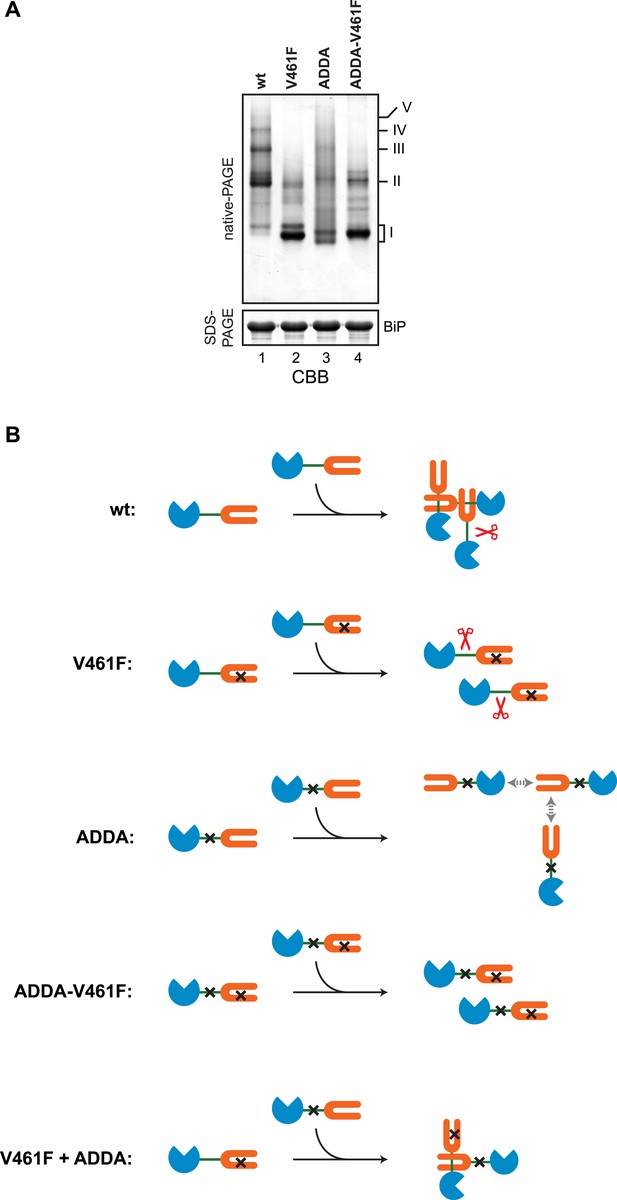
Defective homo-oligomerization of ADDA mutant BiP.
(A) Coomassie-stained native gel of wildtype (wt), V461F, ADDA and compound ADDA-V461F mutant BiP (each at 50 µM) purified from E. coli and allowed to equilibrate 16 hr at room temperature before analysis by native-PAGE (upper panel). The major species have been numbered I-V by order of descending mobility. The same samples were applied to SDS-PAGE as a loading control (lower panel). (B) A schema of the BiP species involved in the experiment in ‘A’ above and in Figure 7. The BiP nucleotide binding domain (NBD) is colored blue, its substrate binding domain (SBD) orange and the interdomain linker green. The V461F mutation (in the SBD) and the ADDA mutation (in the interdomain linker) are indicated by crosses. SubA is cartooned by scissors. BiPV461F is unable to engage in substrate binding and its interdomain linker is unprotected. BiPADDA, which is able to bind substrates is unable to undergo linker-binding mediated homo-oligomerization, and is uncleaved by SubA [the gray double-headed arrows depict putative alternative (weak) substrate binding interactions unmasked by the ADDA mutation]. BiPADDA and BiPV461F are able to form heterodimers, but these are resistant to cleavage by SubA.
BiP oligomeric status responds to changes in ER unfolded protein load.
(A) Immunoblot of endogenous BiP from CHO-K1 cell lysates resolved by native-PAGE. Where indicated the cells were exposed to cycloheximide (CHX, 100 µg/ml), novobiocin (NB, 0.5 mM) or both and the lysate was depleted of ATP by incubation with hexokinase and glucose (+HK). The major species observed on the native gel are numbered by order of descending mobility (I-III) and the ‘B’ form associated with cycloheximide and ‘A’ form present in untreated cells are noted. Immunoblot of the same sample resolved by SDS-PAGE (lower panel) reports on total BiP loaded and on eIF2α as a loading control. (B) As in ‘A’. ATP-depleted lysates of CHO-K1 cells harboring a stable Fv2E-PERK transgene encoding a dimerizer drug (AP20187)-inducible form of the eIF2α-kinase PERK. Where indicated the cells were exposed for 2 hr to AP20187 (15 nM) to activate Fv2E-PERK, in the presence or absence of novobiocin. The lower panels report on the levels of BiP, total eIF2α and phosphorylated eIF2α in the lysates. (C, D) As in ‘A’. Where indicated, cells were treated alone or sequentially with cycloheximide (CHX, 100 µg/ml) and the lumenal calcium depleting agent thapsigargin (Tg, 0.5 µM), the reducing agent DTT (1 mM) that interferes with disulfide bond formation, the glycosylation inhibitor tunicamycin (Tm, 2.5 µg/ml) or the protein misfolding-inducing proline analog, azetidine (Aze, 4 mM) in the indicated order and time. ATP was depleted from all samples during cell lysis.
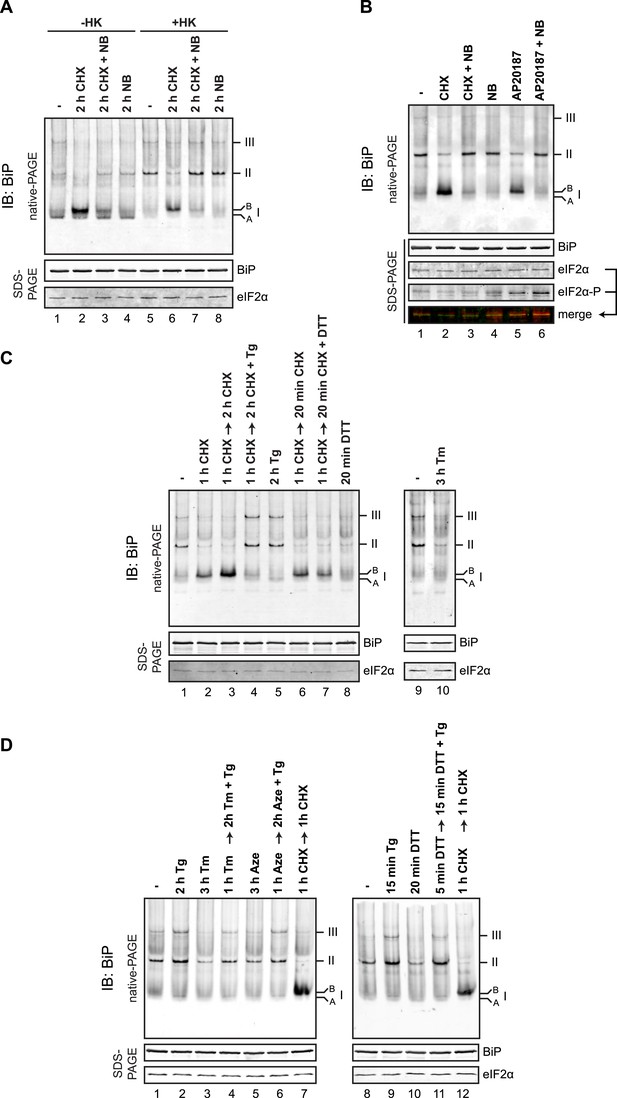
Time-course of evolution of the cycloheximide-dependent ‘B’ form of BiP.
(A) Immunoblot of endogenous BiP from ATP-depleted lysates of CHO-K1 cells resolved by native-PAGE after the indicated time of exposure to cycloheximide (CHX, 100 µg/ml). The major species observed on the native gel have been numbered by order of descending mobility (I-III) and the ‘B’ form associated with cycloheximide and the ‘A’ form observed in untreated cells are noted. Immunoblot of the same samples resolved by SDS-PAGE (lower panel) reports on total BiP loaded and on eIF2α as a loading control. (B) Immunoblot of endogenous BiP from lysates of CHO-K1 resolved by native-PAGE following the indicated pharmacological manipulations, as in Figure 8C [100 µg/ml cycloheximide (CHX), 0.5 µM thapsigargin (Tg), 1 mM DTT]. The lysates were prepared without ATP depletion. Note: In absence of ATP depletion BiP oligomers dissociated into a high-mobility form of species I, ‘A’ form, whose relationship to the ‘B’ form is thereby highlighted. The same samples were applied to SDS-PAGE and BiP and eIF2α were detected as a loading control.
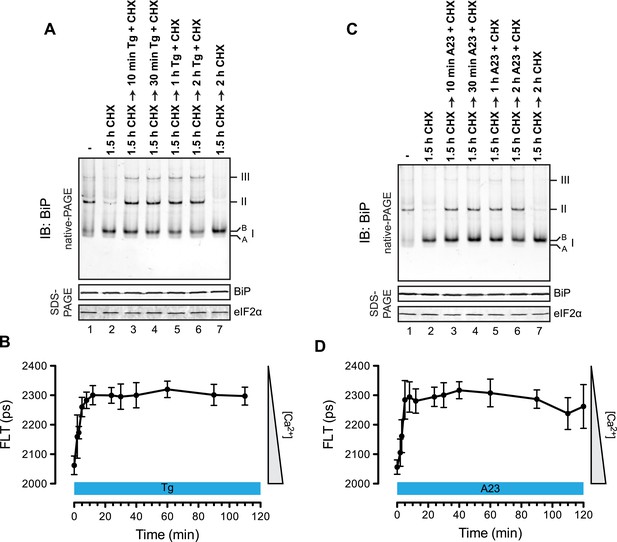
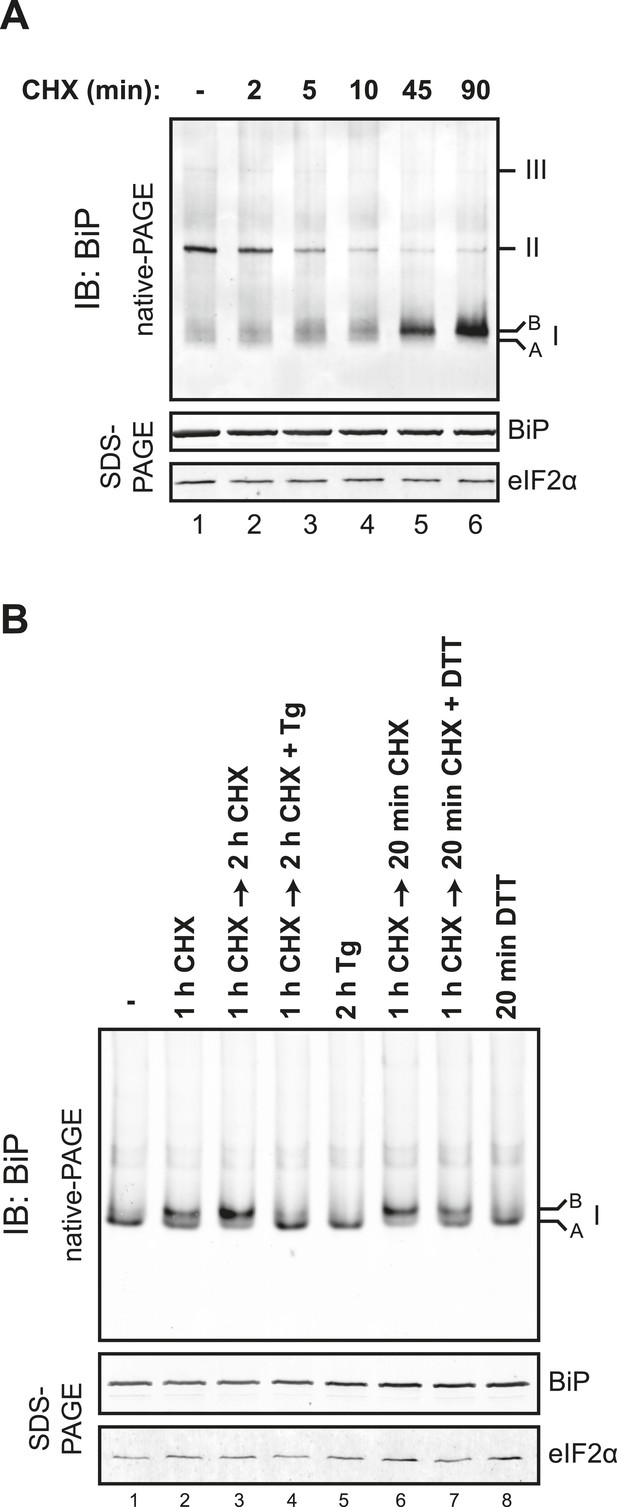
Rapid BiP oligomerization in response to ER calcium depletion.
(A) Immunoblot of endogenous BiP from ATP-depleted lysates of CHO-K1 cells resolved by native-PAGE. Where indicated the cells were exposed to cycloheximide (CHX, 100 µg/ml) followed by thapsigargin (Tg, 0.5 µM). The major species observed on the native gel have been numbered by order of descending mobility (I-III) and the ‘B’ and ‘A’ forms noted. Immunoblot of the same samples resolved by SDS-PAGE (lower panel) reports on total BiP loaded and on eIF2α as a loading control. (B) Plot of time-dependent change of the donor fluorescence lifetime (FLT) of ER localized D1ER cameleon in CHO-K1 cells treated for 1.5 hr with CHX followed by exposure to Tg as in ‘A’. The increase in donor fluorescence lifetime, a consequence of loss of calcium-dependent intra-molecular FRET in cameleon, reports on the kinetics of ER calcium depletion. Bars: mean ± SEM. (C) As in ‘A’, substituting the calcium ionophore A23187 (A23, 10 µM) for thapsigargin. (D) As in ‘B’ substituting the calcium ionophore A23187 (A23, 10 µM) for thapsigargin.
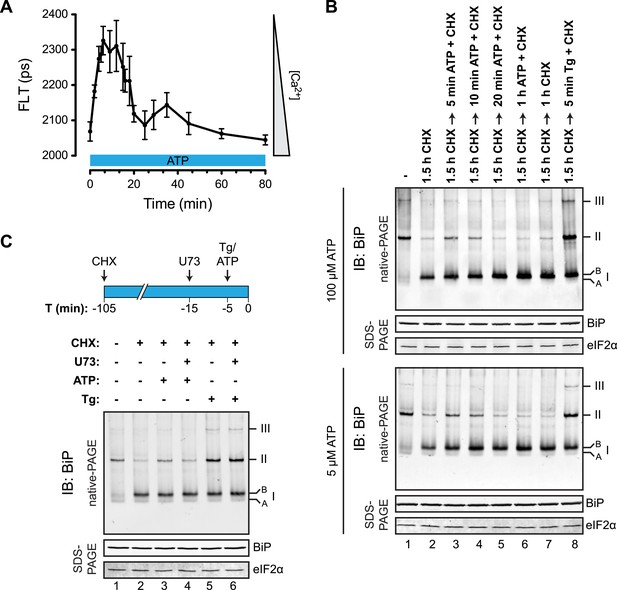
Purinergic receptor engagement promotes BiP oligomerization at physiological levels of ER calcium depletion.
(A) Plot of time-dependent change if the donor fluorescence lifetime of ER localized D1ER cameleon in CHO-K1 cells treated for 1.5 hr with cycloheximide (100 µg/ml) followed by exposure to extracellular ATP (100 µM), a ligand of endogenous purinergic receptors. The increase in donor fluorescence lifetime, a consequence of loss of calcium-dependent intra-molecular FRET in D1ER cameleon, reports on the kinetics of ER calcium depletion, which is notably transient. Bars: mean ± SEM. (B) Immunoblot of endogenous BiP from ATP-depleted lysates of CHO-K1 cells resolved by native-PAGE. The major species observed on the native gel have been numbered by order of descending mobility (I-III) and the ‘A’ and ‘B’ forms are indicated. Where indicated the cells were exposed to cycloheximide (CHX, 100 µg/ml) followed by ATP. Immunoblots of the same samples resolved by SDS-PAGE are shown below the native gels and report on total BiP loaded and on eIF2α as a loading control. (C) Native-PAGE immunoblot of endogenous BiP. As indicated in the schema above, cells were treated sequentially with CHX (100 µg/ml), ATP (100 µM), thapsigargin (Tg, 0.5 µM) or the phospholipase C inhibitor U73122 (U73, 6 µM) in the indicated order. DMSO was used instead of U73 as a vehicle control. Immunoblot of the same samples resolved by SDS-PAGE (lower panel) reports on total BiP loaded and on eIF2α as a loading control.
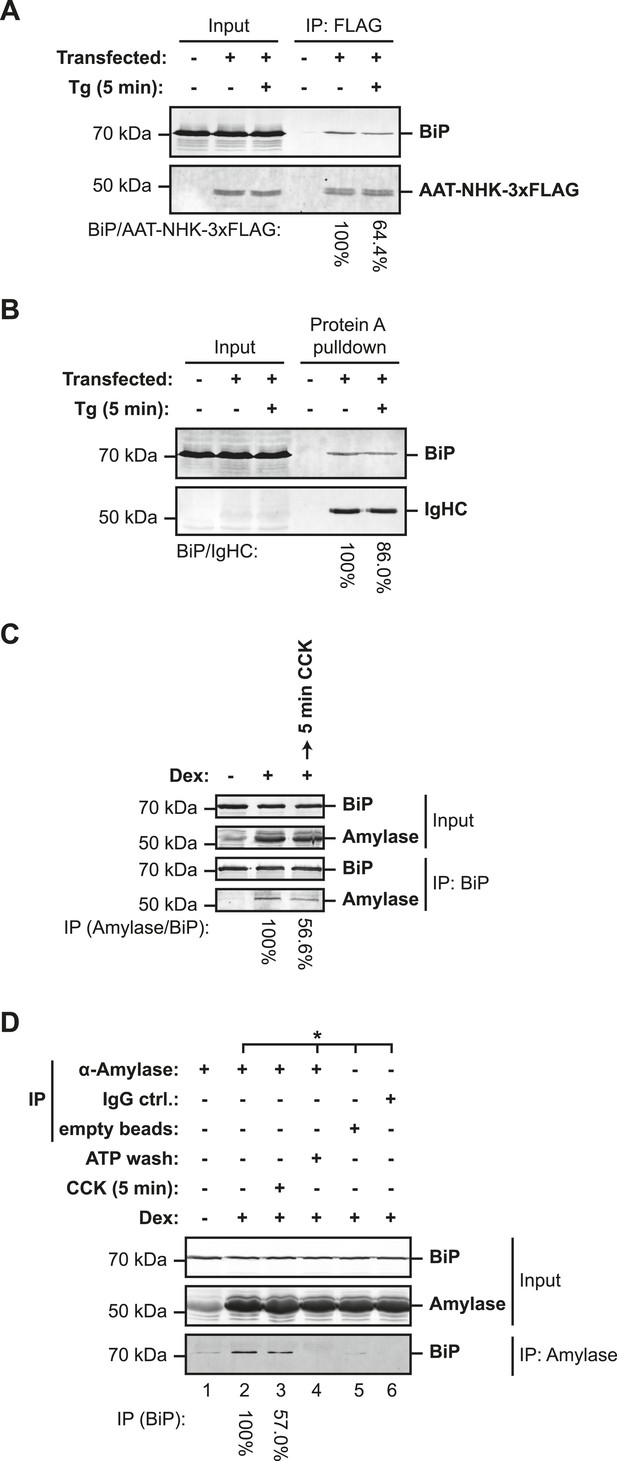
Lumenal calcium depletion-mediated dissociation of BiP from its client proteins.
(A) SDS-PAGE and immunoblot of BiP and FLAG-tagged misfolded null Hong Kong variant of α1-antitrypsin (AAT-NHK-3xFLAG) in ATP-depleted lysates of transfected CHO-K1 cells (Input) or recovered in an anti-FLAG immunoprecipitation. Where indicated the cells were exposed to thapsigargin (Tg, 1 µM) for 5 min before lysis. The recovery of BiP, normalized to the α1-antitrypsin signal in the immunoprecipitation, is noted below each lane and is set to 100% in the untreated sample. Shown is a representative experiment reproduced three times with similar, Tg-mediated disruption of the BiP-AAT complex. (B) SDS-PAGE and immunoblot of BiP and immunoglobulin heavy chain (IgHC) in ATP-depleted lysates of transfected CHO-K1 cells (Input) or recovered in complex with S. aureus protein A. Where indicated the cells were exposed to Tg (1 µM) for 5 min before cell lysis. The recovery of BiP, normalized to the immunoglobulin heavy chain signal in the protein A complex, is noted below each lane and is set to 100% in the untreated sample. Shown is a representative experiment reproduced three times with similar, Tg-mediated disruption of the BiP-IgHC complex. (C) SDS-PAGE and immunoblot of endogenous BiP and amylase in ATP-depleted lysates of AR42j cells (Input) or following immunoprecipitation of BiP from the same (IP). Where indicated the cells were differentiated into an acinar lineage with dexamethasone (Dex, 100 nM) for 48 hr followed by exposure to cholecystokinin (CCK, 4 µM) for 5 min. The recovery of amylase, normalized to the BiP signal in the immunoprecipitation is noted below each lane and is set to 100% in the untreated sample. Shown is a representative experiment reproduced three times with similar, CCK-mediated disruption of the BiP-amylase complex. (D) SDS-PAGE and immunoblot of endogenous BiP and amylase from ATP-depleted lysates of AR42j cells (Input) and recovered in an anti-amylase immunoprecipitation (IP), or two mock IPs. The cells were differentiated into the acinar lineage by dexamethasone (Dex) as in ‘C’ and exposed to cholecystokinin (4 µM), as indicated. The recovery of BiP, normalized to the amylase signal in the immunoprecipitation, is noted below each lane and is set to 100% in the untreated sample. Samples from the same lysate are marked with an asterisk (*). The immunoprecipitated sample in lane 4 was subjected to an ATP elution step, to confirm that BiP associated with amylase through a conventional substrate-binding mechanism. Shown is a representative experiment reproduced three times with similar, CCK-mediated disruption of the amylase–BiP complex.
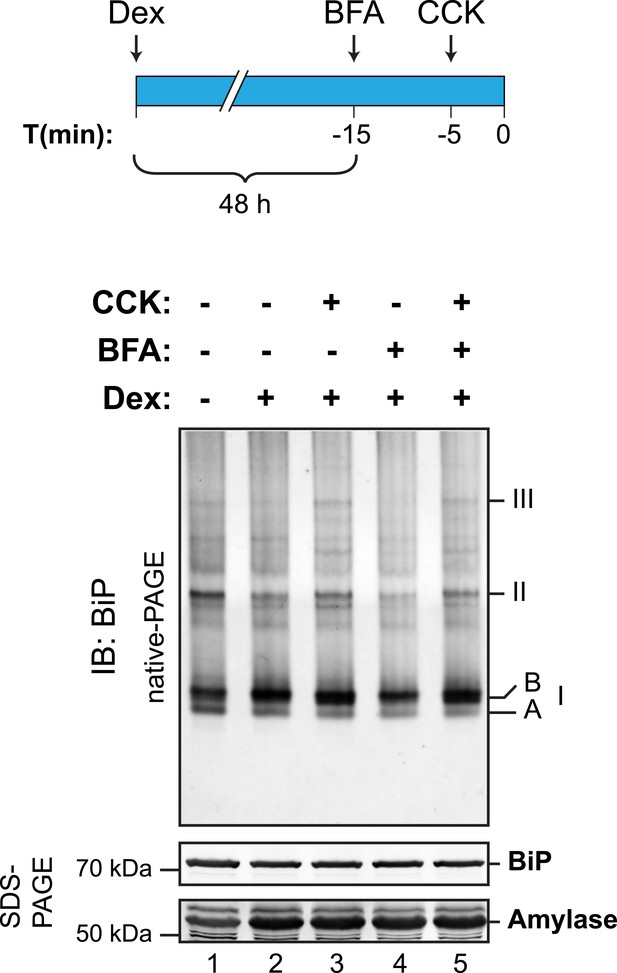
Cholecystokinin-mediated lumenal calcium depletion promotes BiP oligomerization in AR42j cells.
Immunoblot of endogenous BiP from ATP-depleted lysates of AR42j cells resolved by native-PAGE. Where indicated the cells were treated with dexamethasone (Dex, 100 nM, to promote differentiation into secretory cells), cholecystokinin (CCK, 1 µM, to promote ER calcium store depletion) or brefeldin A (BFA, 10 µg/ml, to block export from the ER and promote unfolded protein stress; BFA is particularly effective in this regard in AR42j cells), according to the schedule schematized above. The major species observed on the native gel have been numbered I-III by order of descending mobility, as are the two forms of BiP detected in the higher mobility region of the gel (‘A’ and ‘B’). BiP and amylase immunoblots of the same samples resolved by SDS-PAGE are presented in the panels below. Note that the CCK-mediated increase in abundance of BiP oligomers is highlighted on the background of BFA treatment.
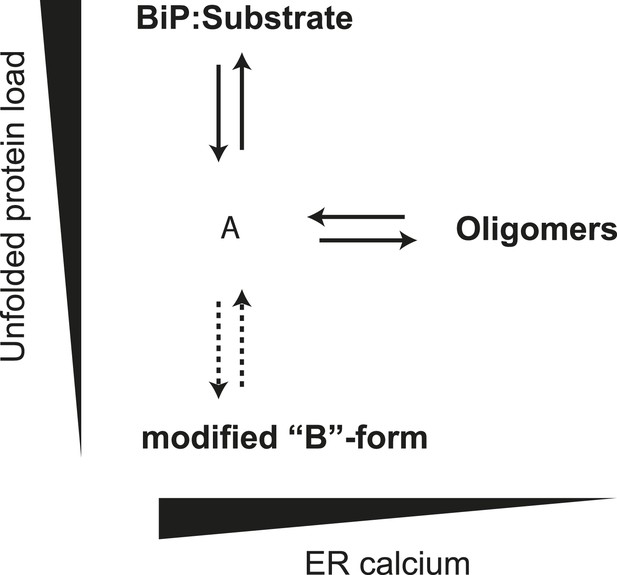
Schema of the proposed relationships between the different forms of BiP.
The monomer (‘A’ form) at the center of the schema binds clients and is shifted to BiP:Substrate complexes by mounting unfolded protein load. With diminishing clients it partitions to inactive oligomers, with whom it exists in equilibrium. This equilibrium is influenced by ER calcium, whose depletion favors the oligomeric form. The ‘A’ form is also in equilibrium with modified BiP (‘B’ form, likely ADP-ribosylated or AMPylated BiP) and this equilibrium too is influenced by unfolded protein burden but on a slower time scale than oligomer formation and disassembly.
Additional files
-
Supplementary file 1
Plasmids used.
- https://doi.org/10.7554/eLife.08961.026





