An ancient Pygo-dependent Wnt enhanceosome integrated by Chip/LDB-SSDP
Figures
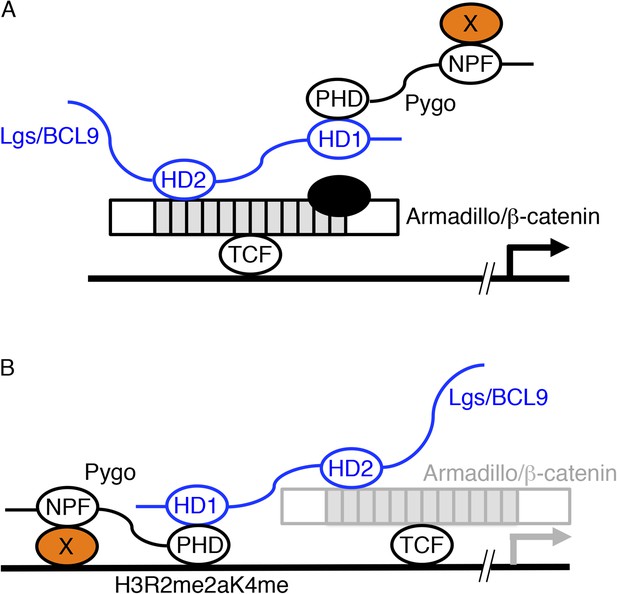
Two models of Pygo function.
(A) The co-activator model (Kramps et al., 2002; Hoffmans et al., 2005): the NPF ligand (X, orange) is a transcriptional co-activator recruited to dTCF enhancers exclusively during Wnt signaling through the Pygo-Legless/BCL9 adaptor chain (Stadeli and Basler, 2005), co-operating with other transcriptional co-activators recruited to the C-terminus of Armadillo (such as chromatin remodelers and modifiers, black) in stimulating Wg-induced transcription. (B) The Armadillo-loading model (Townsley et al., 2004): the NPF ligand (X, orange) mediates constitutive tethering of Pygo to dTCF enhancers prior to Wg signaling, jointly with PHD-mediated recognition of H3K4me1 (marking poised enhancers; Kharchenko et al., 2011) or H3R2me2aK4me1 (marking silenced enhancers in the process of being activated; Kirmizis et al., 2007), priming these enhancers for Wg responses via its ability to capture Armadillo (once available during Wg signaling, indicated by grey) through the Legless/BCL9 adaptor. In both models, the homology domain 1 (HD1) of Lgs/BCL9 binds to the Pygo PHD finger, while HD2 binds to the N-terminus of the Armadillo Repeat Domain (light grey) of Armadillo/β-catenin (Kramps et al., 2002; Sampietro et al., 2006; Fiedler et al., 2008; Miller et al., 2013).
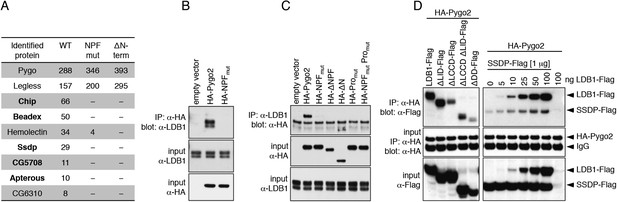
Pygo NPF binds to ChiLS.
(A) Top proteins associated with wt but not NPF-mutant Pygo in S2 cells (unweighted spectral counts >95% probability are given); bold, ChiLS and its ligands. (B–D) Western blots of coIPs from transfected HEK293T cells, showing NPF-dependent coIP of (B, C) endogenous LDB1 with HA-Pygo2 or (D) wt vs truncated LDB1-Flag with HA-Pygo2 (left), and LDB1-Flag +/− SSDP-Flag with HA-Pygo2 (right; increasing amounts of LDB1-Flag indicated above panel).
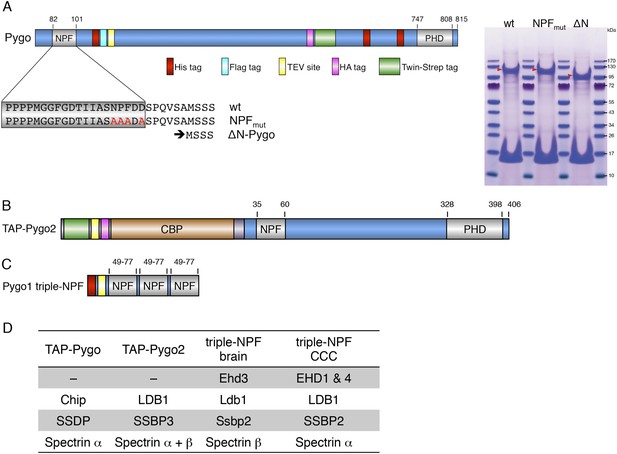
Baits used for mass spectrometry analysis.
(A) Cartoon of tandem-tagged Drosophila Pygo bait, generated for expression in stably transfected S2 cells (continuously selecting with 5 μg ml−1 puromycin), and subsequent tandem purification (TAP) of associated proteins using α-Flag followed by Strep-tactin pull-down; red arrowheads indicate wt and mutant Pygo baits after electrophoresis on a stained polyacrylamide gel prior to excision for mass spectrometry analysis. (B) Cartoon of tandem-tagged human Pygo2 bait (subcloned in V51 pGLUE), for expression in stably transfected HEK293T cells (continuously selecting with 2 μg ml−1 puromycin), for TAP purification of associated proteins with streptavidin followed by Calmodulin resin, essentially adopting a protocol previously described (Angers et al., 2006). (C) Cartoon of triple-NPF bait from mouse Pygo1, generated by trimerizing an extended NPF motif (amino acids 49-77), or a corresponding NPF > AAA triple mutant, for Ni-NTA pull-down experiments with lysates from fresh mouse brains (strain C57Bl6J), or from SW480 or COLO320 colorectal cancer cell (CCC) lines (obtained from Marc de la Roche; de la Roche et al., 2014). Interacting proteins were eluted with 6M urea, and samples were prepared for analysis using a filter-aided sample preparation method, essentially as described (Wisniewski et al., 2009). (D) Hits identified with TAP-Pygo and TAP-Pygo2 baits in both S2 and HEK293T cells, respectively (see A, B), and overlap with hits identified by triple-NPF immunoprecipitations from mouse brain and CCC lysates (see C), which include ChiLS components. Note that EHD paralogs were only found in the lysate-based approaches (amongst the top hits, due to the cytoplasmic abundance of these proteins) but not with full-length Pygo or Pygo2; spectrins were also commonly found, but the significance of this is unclear.
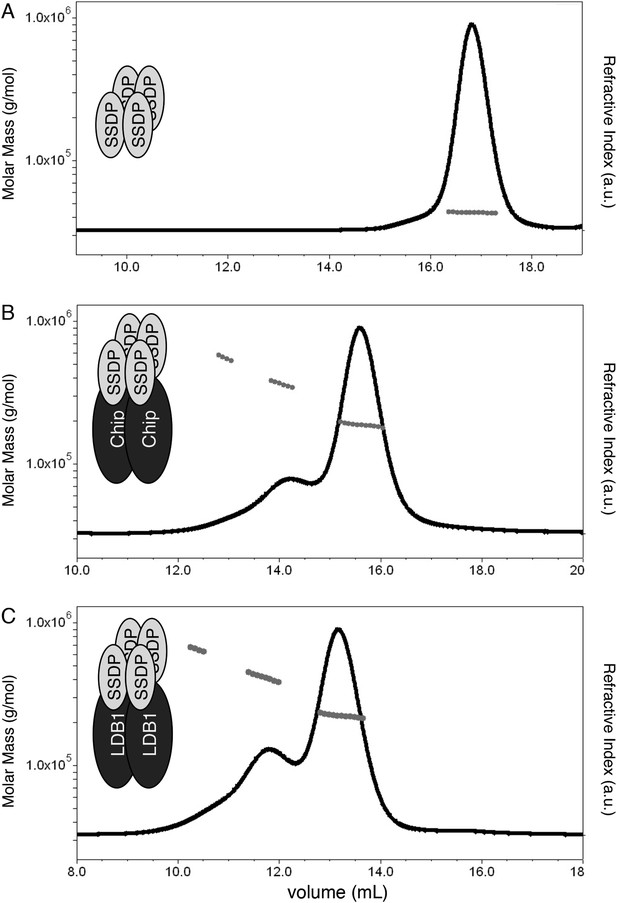
Stoichiometry of the ChiLS complex.
SEC-MALS of (A) Lip-SSDP1-92, or co-expressed (B) Lip-SSDP1-92 + MBP-Chip205-436 or (C) Lip-SSDP1-92 + MBP-LDB156-285; solid black lines, elution profile as detected by the RI detector; grey circles, molecular mass; cartoons in panels indicate stoichiometries consistent with the measured molecular masses.
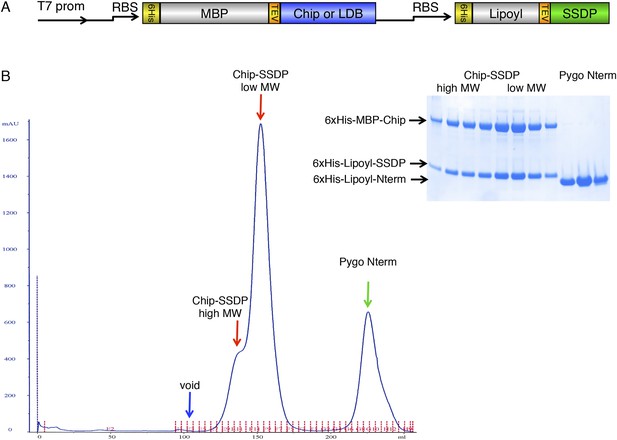
Gel filtration of ChiLS.
(A) Bi-cistronic expression vector (with T7 promoter) used for bacterial expression of ChiLS complex, encoding 6xHis-MBP-Chip205-436 or 6xHis-MBP-LDB156-285 followed by 6xHis-Lipoyl-SSDP1-92 with its own ribosome-binding site (RBS). (B) Elution profile of MBP-Chip205-436, Lipoyl-SSDP1-92 (after co-expression with vector shown in A) and incubation with excess 6xHis-Lipoyl-Pygo67-107 (Pygo Nterm), revealing three main species, as indicated above graph, corresponding to free Pygo Nterm, Chip-SSDPlow and Chip-SSDPhigh complexes, as shown by polyacrylamide gel electrophoresis (inset); note that the high-molecular weight shoulder (Chip-SSDPhigh) corresponds to a dimer of the major ∼190 kDa low-molecular weight species (Chip-SSDPlow), whereby the latter corresponds to an MBP-Chip dimer bound to an SSDP tetramer (see Figure 3B). Chip-SSDPlow and Chip-SSDPhigh complexes cannot be further separated by subsequent anion exchange chromatography, and do not seem to interconvert, as judged by iterative G-200 gel chromatography. Both complexes bind to 15N-Pygo-NPF (see Figure 4), however, Pygo Nterm dissociates from ChiLS during gel filtration (see inset), indicating a low affinity to ChiLS (estimated to be mid-μM).
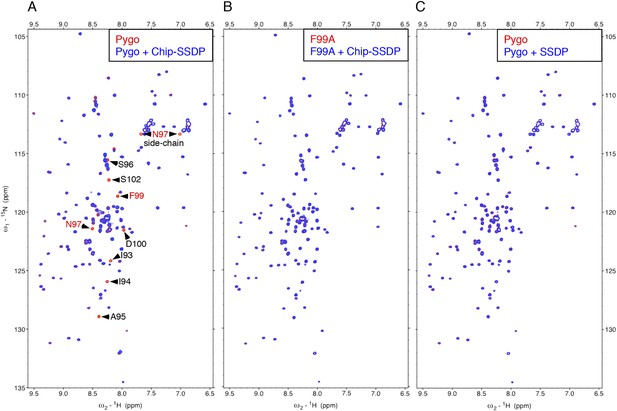
Direct NPF-dependent binding of Pygo by ChiLS.
Overlays of HSQC spectra of 50 μM 15N-labeled wt or F99A mutant Pygo67-107 alone (red) or probed with (A, B) MBP-Chip205-436- Lip-SSDP1-92 or (C) Lip-SSDP1-92 alone (blue); interacting residues are labeled, with NPF in red (binding to P is not detectable by HSQCs). The HSQC obtained with 50 μM of minimal 15N-labeled Pygo87-102 is indistinguishable from that shown in (A).
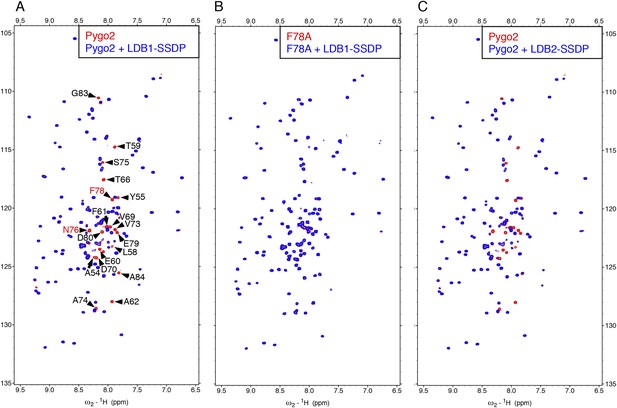
Direct NPF-dependent binding of Pygo2 by ChiLS.
Overlays of BEST-TROSY spectra of 50 μM 15N-labeled (A, C) wt or (B) F78A mutant human Pygo237-93 alone (red) or probed with 100 μM (A, B) MBP-LDB156-285- Lipoyl-SSDP1-92 (blue) or (C) MBP-LDB220-246- Lipoyl-SSDP1-92 (blue); interacting residues are labeled, with NPF motif in red (binding to P is not detectable by BEST-TROSY).
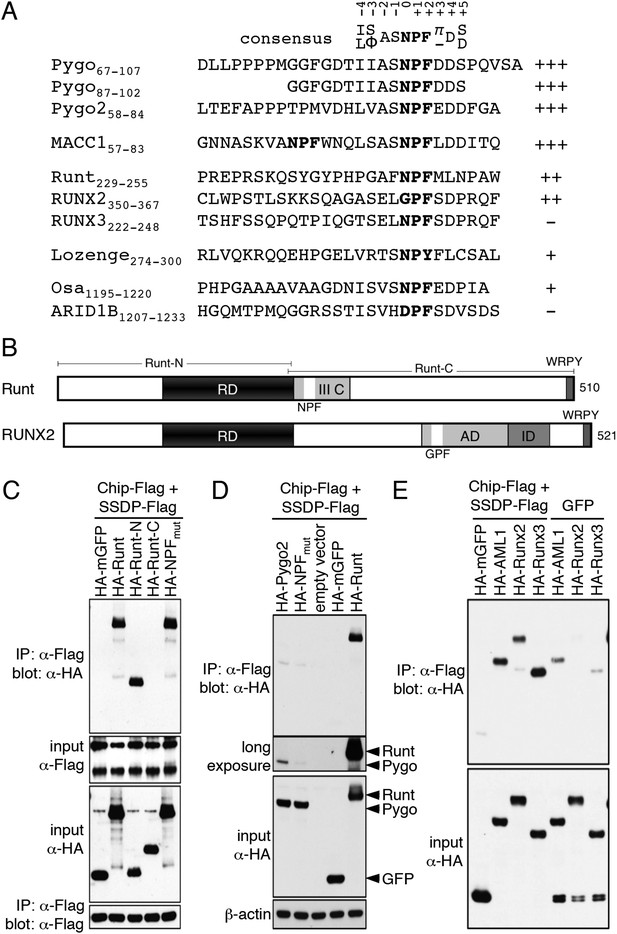
ChiLS binds to RUNX NPFs.
(A) Summary of NMR binding assays of 15N-labeled NPF fragments probed with ChiLS; +–+++, estimates of binding affinities, based on minimal ChiLS concentrations required for line-broadening; −, no binding (see also Figure 4); top, preferred NPF context in strong binders (numbering of positions as in de Beer et al., 2000). (B) Schematic of RUNX orthologs, with DNA-binding domain (RD, black), region IIIC, activation and inhibitory domains (AD, ID), NPF (or GPF) and WRPY indicated. (C–E) Western blots as in Figure 2, showing coIP between co-expressed proteins as indicated above panels; for Runt-N and Runt-C, see (B); mGFP (control).
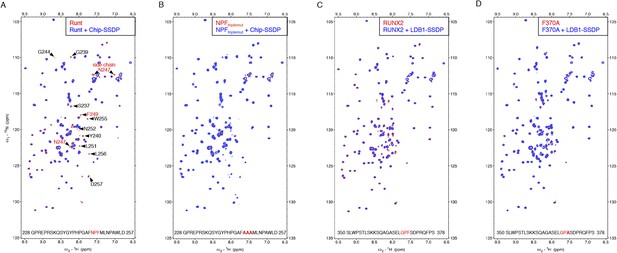
Specific recognition of RUNX NPFs by ChiLS.
(A, B) Overlays of HSQC spectra of 50 μM 15N-labeled 6xHis-Lipoyl-tagged (A) wt or (B) NPF > AAA triple-mutant Runt-NPF228-257 alone (red) or probed with 300 μM MBP-Chip205-436-Lipoyl-SSDP1-92 (blue), as indicated in panels; interacting residues are labeled in (A). (C, D) Overlays of HSQC spectra of 50 μM 15N-labeled 6xHis-Lipoyl-tagged (C) wt or (D) F > A mutant RUNX2-NPF350-378 alone (red) or probed with 300 μM MBP-LDB156-285- Lipoyl-SSDP1-92 (blue), as indicated in panels. Note that a triple-NPF mutation (NPF > AAA) is required to block binding of Runt to ChiLS (B) while a single F > A mutant Runt still binds to ChiLS (at this concentration). Sequences of 15N-labeled NPF peptides are given in the panels (red, NPF motifs; bold, mutated residues). RUNX3 binding was undetectable with NPF-containing peptides from human RUNX3 (amino acids 227–249; SQPQTPIQGTSELNPFSDPRQFD) or mouse Runx3 (amino acids 214-269).
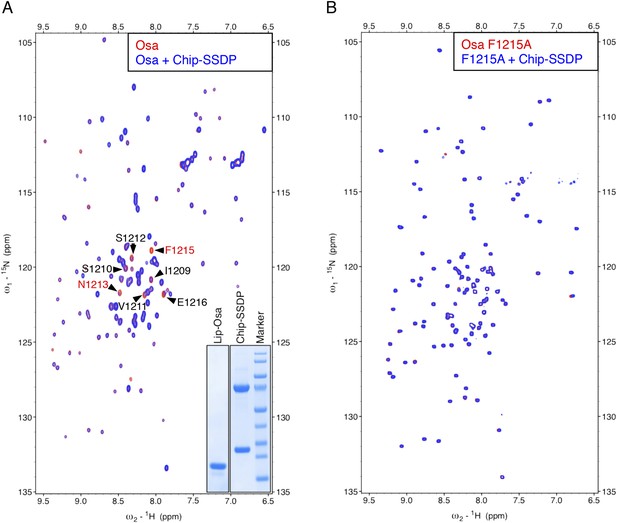
Specific recognition of Osa NPF by ChiLS.
Overlays of (A) HSQC or (B) BEST-TROSY spectra of 50 μM 15N-labeled 6xHis-Lipoyl-tagged (A) wt or (B) F1215A mutant Osa1195-1221 alone (red), or probed with 300 μM MBP-Chip205-436- Lipoyl-SSDP1-92 (blue), as indicated in panels; interacting residues are labeled in (A). Inset in (A) shows purified 6xHis-Lipoyl-Osa1195-1220 and MBP-Chip205-436- Lipoyl-SSDP1-92 complex separated by polyacrylamide gel electrophoresis. Note also that the NPF-containing peptide we tested for ARID1B was negative, however, this large protein contains further matches to the NPF consensus sequence that might bind ChiLS (to be tested in future studies). The same applies for its ARID1A paralog.
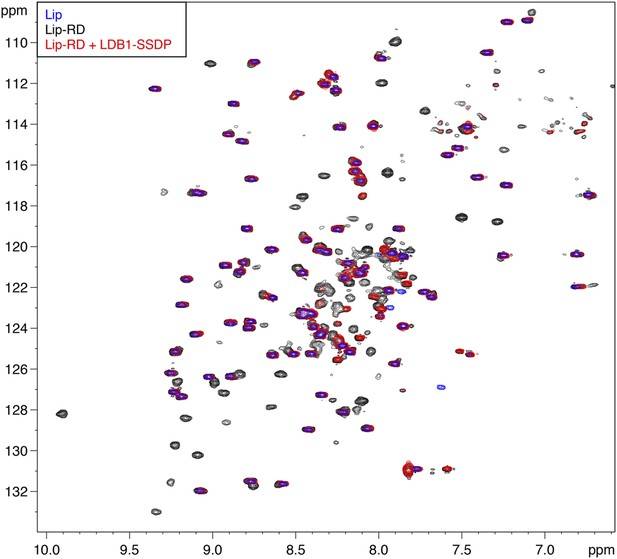
Direct binding of RUNX RD by ChiLS.
Overlays of BEST-TROSY spectra of 15N-labeled 6xHis-Lipoyl (blue), 6xHis-Lipoyl-RUNX2-RD102-234 (black) and 6xHis-Lipoyl-RUNX2-RD102-234 + 300 μM MBP-LDB156-285- Lipoyl-SSDP1-92 (red), as indicated in panels. Note that the resonances affected by incubation with LDB1-SSDP are predominantly RD-specific (black) whereas most Lip residues (marked by blue) are unaffected.
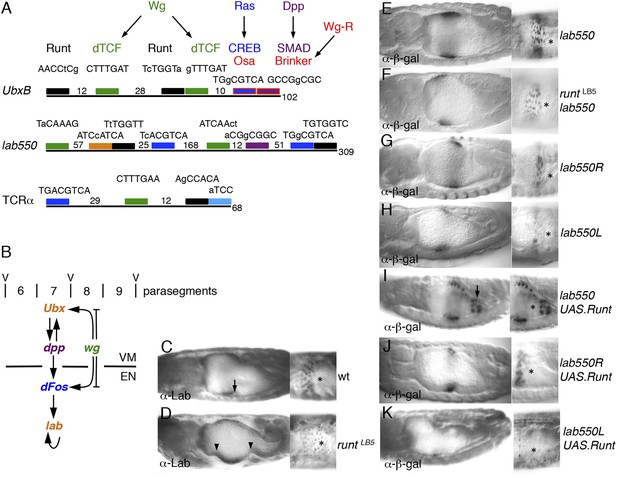
Runt acts through the labial midgut enhancer.
(A) Cartoon of UbxB, lab550 and TCRα enhancers, with the following binding sites (responding to signals, as indicated in brackets): green, dTCF (Wg); purple, SMAD (Dpp); light-blue, Ets (Ras); blue, CRE (Ras); in UbxB, the SMAD binding site also mediates Wg-mediated repression by Brinker (red), and CRE mediates Osa-mediated repression (see text); black, RUNX; orange, Labial; residue numbers between binding sites are given (right, total length; note that UbxB and lab550 extend beyond these modules which however contain all known functional binding sites); capital letters, matches to binding site consensus sequences (C/G G C/G G G T C/G for RUNX; Melnikova et al., 1993). (B) Cartoon of endoderm induction, color-coded as in (A); V, midgut constrictions at parasegment boundaries. (C, D) 14 hr old embryos stained with α-Labial; arrow marks incipient second midgut constriction, lacking in runt mutants (which only form first and third constrictions, arrowheads). (E–K) 12–14 hr old embryos bearing wt or mutant lab550 as indicated on the right, stained with α-β-galactosidase; high magnification views are imaged at different focal planes, to highlight Wg-dependent expression gradients (Wg sources indicated by asterisks).
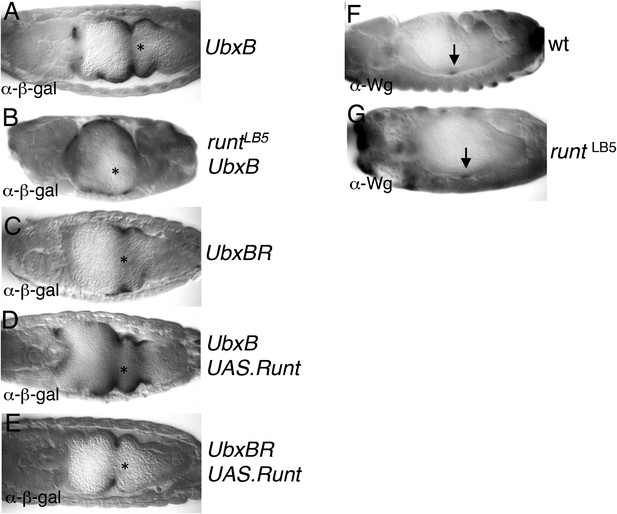
Runt acts through the Ubx midgut enhancer.
(A–E) 14–15 hr old wt or runt mutant embryos, bearing wt or mutant UbxB enhancers (Thuringer et al., 1993), with or without mesodermal GAL4-mediated overexpression of Runt (using 24B.GAL4) as indicated in panels, fixed and stained with α-β-galactosidase (α-β-gal) antibody; Wg sources (in parasegment 8 of the visceral mesoderm; see Figure 6B) are indicated by asterisks. The expression of the mutant enhancer (UbxBR) is reduced, and seen in fewer cells (C), similarly to the expression of the wt enhancer in runt mutant embryos (B), as in the case of lab550 (Figure 6F,G,J), although in both cases, mutating the Runt binding sites has a somewhat stronger effect on expression than loss of Runt (possibly because of a maternal contribution in the runt mutant embryos, which is difficult to rule out). Note also that this minimal enhancer mediates expression posteriorly to parasegment 7 (the Ubx expression domain in the visceral mesoderm), partially escaping repression by high Wg signaling levels near the Wg source (Yu et al., 1998), which depends on displacement of SMAD by the Groucho-recruiting Brinker co-repressor (Saller and Bienz, 2001). (F, G) ∼12 hr old wt or runt mutant embryos, fixed and stained with α-Wg antibody; Wg sources in the visceral mesoderm are indicated by arrows.
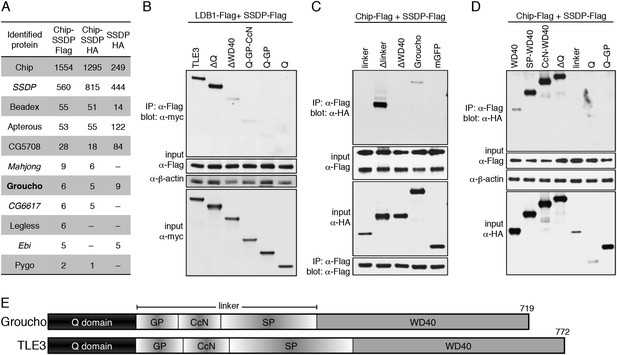
Groucho binds to ChiLS.
(A) Top proteins associated with >2 baits (as indicated in table) in S2 cells, in addition to Legless and Pygo (unweighted spectral counts as in Figure 2A). (B–D) Western blots as in Figure 5C, showing coIP between co-expressed wt and truncated proteins as indicated above panels. (E) Cartoon of Groucho and TLE3, with domains indicated (GP, CcN, SP, semi-conserved elements within linker).
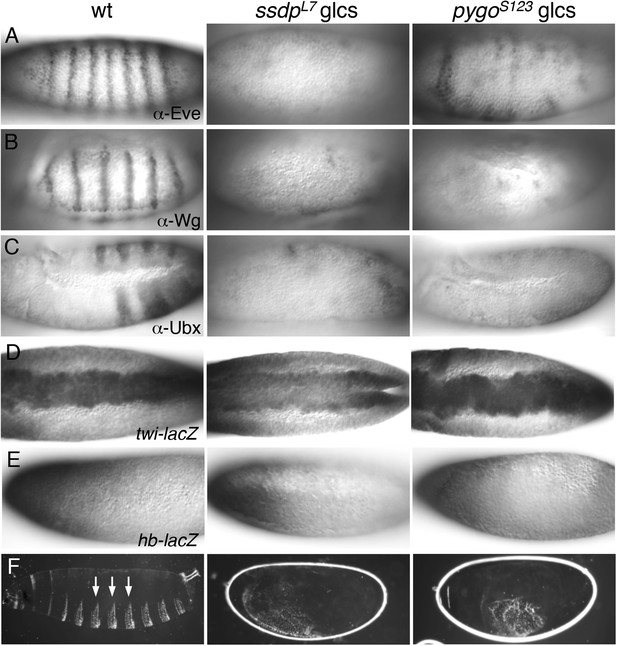
ssdp and pygo mutants show similar early embryonic defects.
(A–E) 3–5 hr old wt and mutant glc embryos as indicated above panels, stained with antibodies as indicated in panels. (F) Larval cuticles of wt and mutant glc embryos; denticle belts (arrows) signify lack of Wg signaling.
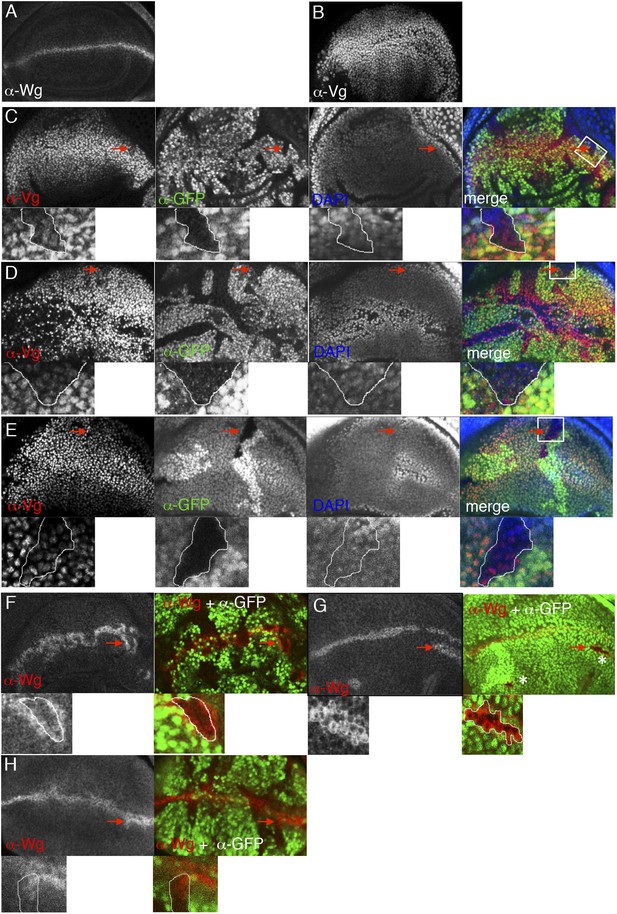
ChiLS is required for Wg and Notch responses in wing discs.
(A, B) Single confocal sections through third larval instar wing discs, fixed and stained with α-Wg or α-Vg antibody, as indicated in panels. wg is activated by Notch signaling along the prospective anterior margin of the third instar disc, while vg is expressed in a Wg-dependent fashion in a broad zone straddling Wg expression along the prospective margin and sustains proliferation in the prospective wing blade (Couso et al., 1995; Rulifson and Blair, 1995; Kim et al., 1996; Klein and Arias, 1999; Micchelli et al., 1997). (C–E) Single confocal sections of wing discs as in (A, B), bearing (C) pygoS123, (D) ssdpL7 or (E) chipe55 mutant clones, stained with α-Vestigial (Vg) (red in merge); clones are marked by absence of GFP (green in merge); all discs were counterstained with DAPI (blue in merge), to label the nuclei (as internal control for the focal plane). Reduction of Vg expression within mutant clones of all three genes is best detectable in the ventral compartment (without Apterous expression) at a distance from the Wg source, as shown in high-magnification insets below panels (from marked squares, marked in merges; clones demarcated by white lines), as previously reported for dTCF mutant clones that are unable to respond to Wg signaling (Schweizer et al., 2003). (F–H) Single confocal sections of wing discs as in (C–E), bearing (F) ssdpL7, (G) chipe55 or (H) pygoS123 mutant clones, stained with α-Wg (red in merge); clones are marked by absence of GFP (green in merge), and discs were counterstained with DAPI; insets below panels are high-magnification views of selected clones (marked by squares in merges) revealing ectopic activation of Wg along the edge of the clone (signifying activation by Notch signaling derived from adjacent wt cells, combined with loss of Wg-mediated repression within the clone; Rulifson and Blair, 1995; Rulifson et al., 1996). Note also that, in the dorsal disc (the Apterous territory), chipe55 mutant clones barely survive (indicated by asterisks in the merge in G), compared to ssdpL7 mutant clones that appear to grow normally, like pygoS123 mutant clones; however, in the absence of Apterous, i.e. in the ventral compartment, chipe55 mutant clones grow normally (see clone marked by arrow in E). Collectively, our clonal analysis indicates that ChiLS is required for Wg and Notch responses in the prospective wing blade and margin, consistent with previous results (Morcillo et al., 1996; Morcillo et al., 1997; van Meyel et al., 2003) similarly to Pygo (Belenkaya et al., 2002; Parker et al., 2002; Thompson et al., 2002), although Chip exhibits a far more stringent requirement for proliferation in the Apterous territory than either SSDP or Pygo.
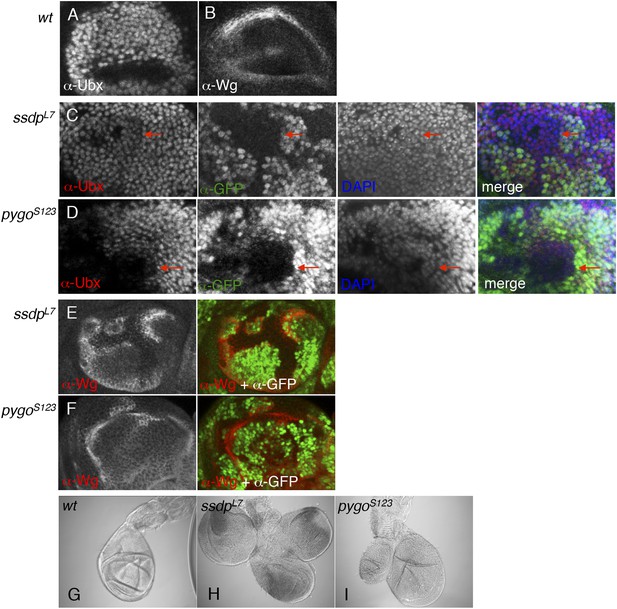
ChiLS is required for Ubx and Wg expression in haltere discs.
(A, B) Single confocal sections through third larval instar haltere discs, fixed and stained with α-Ubx or α-Wg antibody, as indicated in panels. Ubx specifies haltere development by attenuating vg and proliferation in haltere discs, partly through interaction with Wg (Prasad et al., 2003). (C, D) Single confocal sections of haltere discs as in (A, B), bearing pygoS123 or ssdpL7 mutant clones as indicated in panels, stained with α-Ubx antibody (red in merge); clones are marked by absence of GFP (green in merge); all discs were counterstained with DAPI (blue in merge), to label the nuclei (as internal control for the focal plane). Note the reduction of Ubx expression in clones marked by red arrows, which results in slight overproliferation and outgrowth of clone (as judged by the bulging out from the focal plane of the mutant epithelium). (E, F) Single confocal sections of clone-bearing haltere discs as in (C, D), stained with α-Wg antibody (red in merge); clones are marked by absence of GFP (green in merge), and counterstained with DAPI. Note the ectopic expression of Wg along the edges within the mutant clones, similarly to the clones shown in Figure 8—figure supplement 1 (F–H), likely as a result of the same Notch-stimulatory and Wg autoinhibitory and inputs. (G–I) Patterning defects in halteres, reflecting overgrowth of pygoS123 or ssdpL7 mutant clones due to reduced Ubx and ectopic Wg expression in the disc.
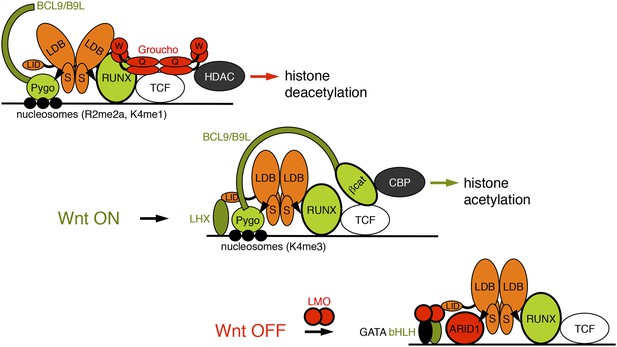
Model of the Wnt enhanceosome.
Switching of the Wnt enhanceosome from OFF (top) to ON (middle), and towards re-repression (bottom), prefacing re-recruitment of Groucho/TLE; S, SSDP; Q, Q domain; W, WD40 domain; NPF-mediated interactions are indicated by arrowheads; green, positively-acting components; red, negatively-acting components; known oncogenes and tumor suppressors are circled in bold. The stoichiometry of ChiLS allows simultaneous interaction with NPF and LID-binding factors, although these may also displace each other, as indicated (except for RUNX, arbitrarily shown as ChiLS-associated throughout). Indicated conformational changes are entirely hypothetical.





