A simple method for generating high-resolution maps of genome-wide protein binding
Figures
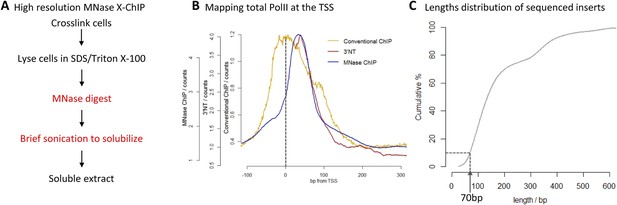
High-resolution X-ChIP-seq of PolII at transcriptional start sites (TSS).
(A) Experimental workflow using MNase to fragment the chromatin. (B) Average PolII profile across TSS in Drosophila. S2 cells as measured by conventional ChIP (Core et al., 2012), high-resolution X-ChIP-seq (fragment lengths 20–70 bp) and 3′NT that maps the position of the polymerase active site via the last ribonucleotide incorporated into the nascent chain (Weber et al., 2014). With 3′NT, the RNA has to be transcribed at least 25 nt in length to be mapped. (C) Length distribution of the mapped paired-end reads from a total PolII high-resolution X-ChIP-seq experiment (Skene et al., 2014).
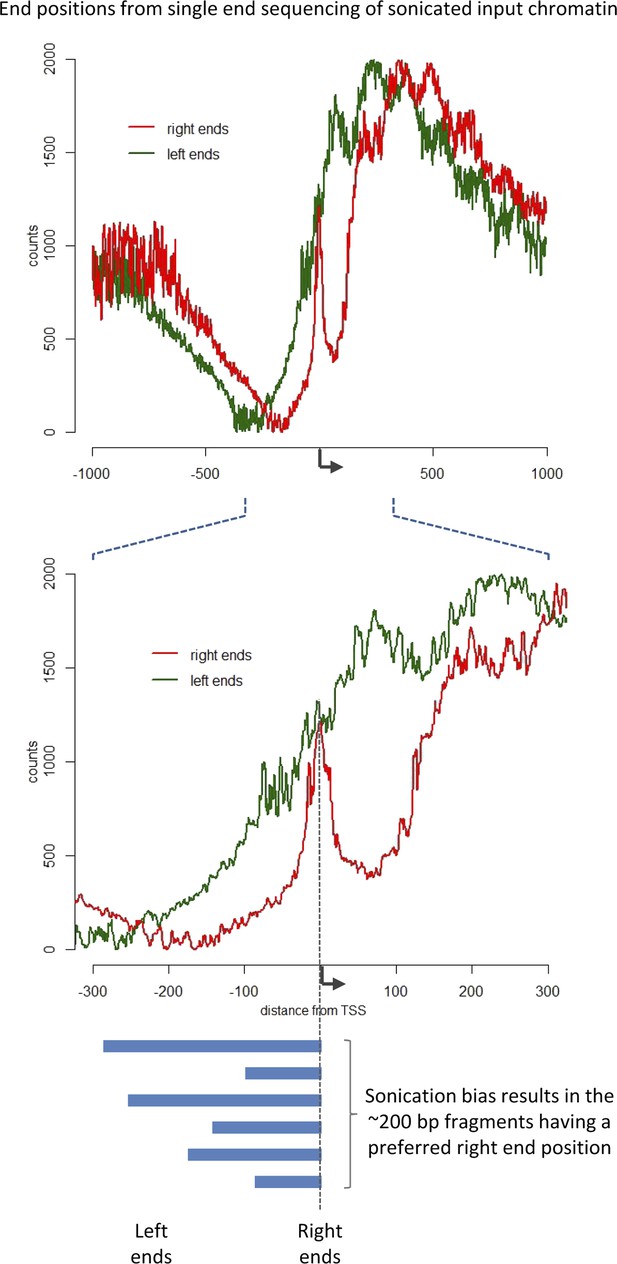
Sonication bias at promoter regions.
The left ends (5′ position on forward strand reads) and right ends (3′ position on reverse strand reads) of fragments from single-end sequencing data of sonicated input chromatin was aligned to the TSS (reads corresponding to genes on the reverse strand were flipped). The non-uniform distribution indicates a bias with the distinct peak at the TSS arising from non-random chromatin fragmentation, with a high probability of having a right end just downstream of the TSS. The input data set from modENCODE ID#3953 (GSE47229) was used. Due to the sonication bias, it is clear that the sonicated fragments do not represent the minimally protected DNA footprint, and as such the length of the recovered DNA fragments provides no information.
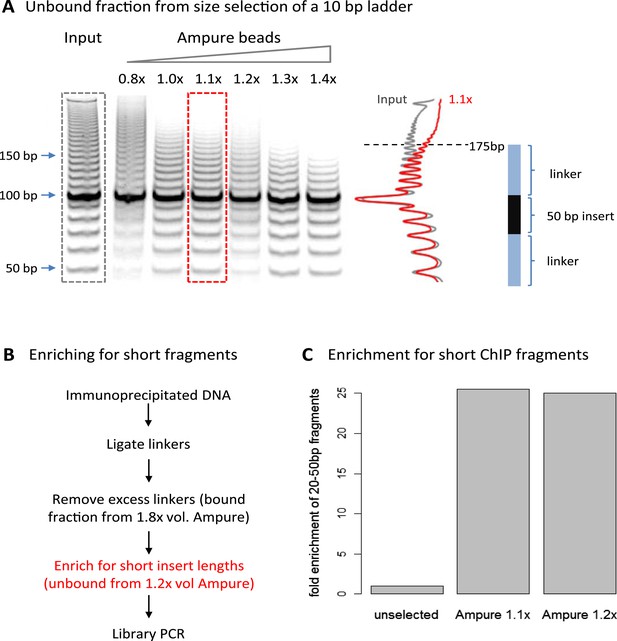
Size selection to enrich for short immunoprecipitated fragments.
(A) Volumetric ratio of AMpure beads to DNA was optimized to selectively retain fragments below 200 bp in the unbound fraction using a 10-bp ladder as a test case. The cartoon indicates the size of ligated product containing a 50-bp insert. (B) Library preparation workflow to enrich for short insert sizes between the ligated linkers. (C) Fold enrichment of sequenced ChIP fragments less than 50 bp after the AMpure size selection as depicted in panel B. This method of enriching for short fragments is specifically applicable to high-resolution X-ChIP-seq, where MNase has been used to generate minimally protected DNA footprints. In contrast, in conventional ChIP-seq where sonication is used, the enrichment of short size classes would be inappropriate as typically fragments are between 200 and 500 bp in length and even extensive sonication can only further fragment chromatin to an average size of 200 bp (Fan et al., 2008).
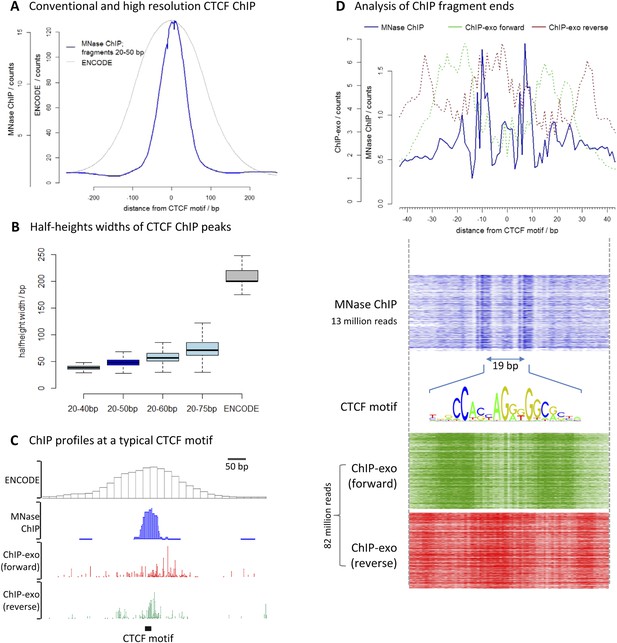
High-resolution X-ChIP-seq provides base-pair resolution of the minimal CTCF sequence motif.
(A) Average CTCF profile at DNase I sites that contain the motif in K562 cells, as measured by conventional ChIP in the ENCODE project and high-resolution X-ChIP-seq (20–50 bp fragments). Sites were determined by identifying the DNase I sites common to K562 and HeLa cells, as defined by the ENCODE project, that contained the 19 bp CTCF consensus binding motif (MA0139.1) by using FIMO analysis with a false discovery rate of 0.01 (Grant et al., 2011). This identified 9403 such 19 bp CTCF motifs within DNase I sites that were at least 500 bp apart. (B) Box plots indicating half-height widths of ChIP peaks at each individual CTCF motif for different size classes of immunoprecipitated fragments in high-resolution X-ChIP-seq and conventional ChIP. (C) ChIP profiles at a typical CTCF motif. For ChIP-exo, the 5′ ends of forward and reverse strands are plotted. (D) The upper graph displays the average profile mapping the position of both of the ends of paired-end reads for the 20–50 bp immunoprecipitated CTCF fragments in high-resolution X-ChIP-seq centered over the CTCF motif. For comparison, the ends for the forward and reverse strands are shown for ChIP-exo. The heatmaps below show the signal ±40 bp for each CTCF motif (defined as CTCF motifs with DNase I sites in both HeLa and K652 cells; n = 9403). The 19 bp between the identified peaks is highlighted and the 19 bp CTCF motif indicated.
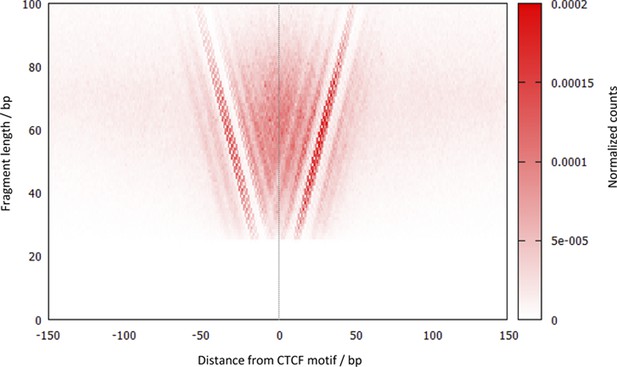
V plot of DNA fragments recovered by CTCF high-resolution X-ChIP-seq.
The V-plot is a midpoint-vs-length map centered on the aligned CTCF motifs (n = 9403), where a dot is placed on a 2D map with the x-axis representing the midpoint position of each immunoprecipitated fragment and the y-axis representing the length of that DNA fragment (Henikoff et al., 2011). Normalized counts represent the number of fragments at each pixel position relative to the total number of pixels. Note that the shorter DNA fragments are more tightly grouped and closely centered over the CTCF motif.
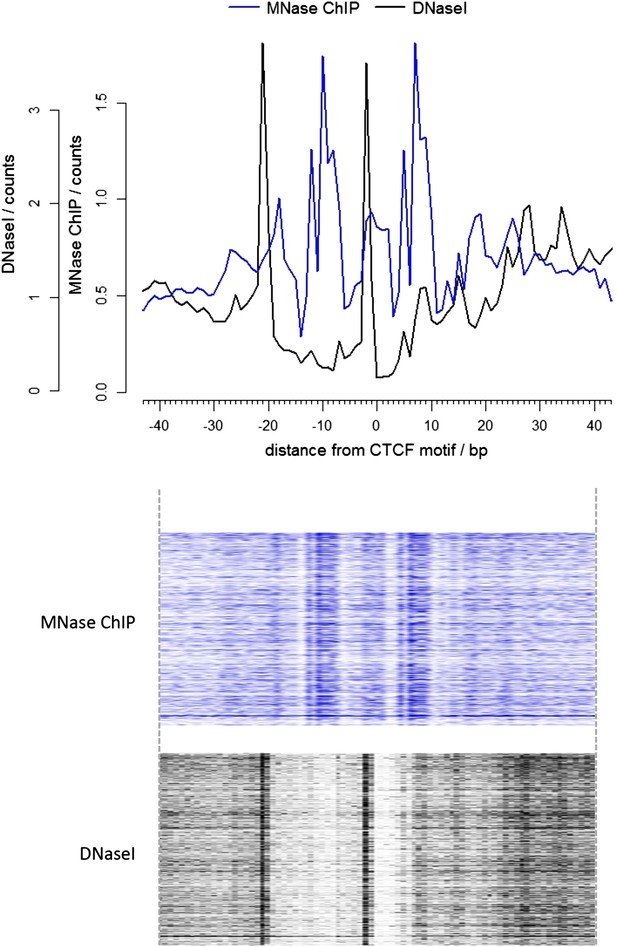
Comparison of the ends of DNA fragments from DNase I and high-resolution X-ChIP-seq centered over the CTCF motif.
The upper graph shows the average profile, and the heatmaps below show the signal ±40 bp for each CTCF motif. The leftward shift of the DNase I footprint likely reflects differences in the steric interactions between CTCF and DNase I and that of MNase.
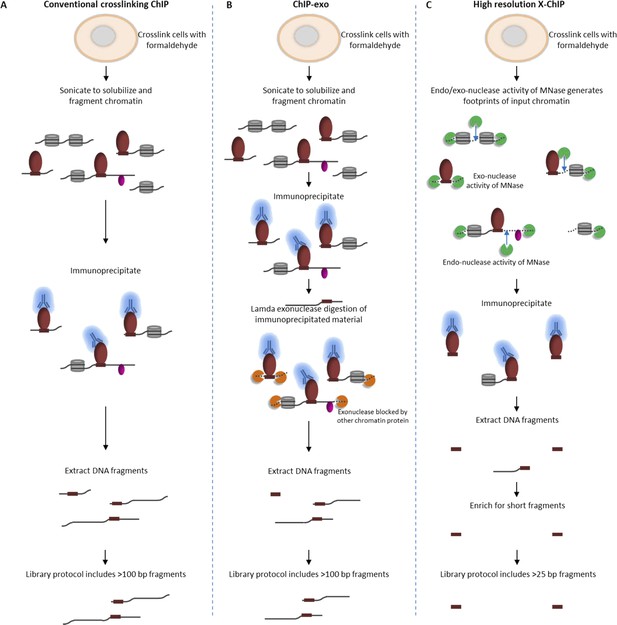
Comparison of different ChIP methodologies and how the resolution obtained depends on the fragmentation strategy used.
The fragmentation strategy is shown for (A) conventional ChIP-seq, (B) ChIP-exo and (C) high-resolution X-ChIP-seq. In high-resolution X-ChIP-seq, MNase generates minimally protected DNA fragments that are represented by the lengths of the extracted DNA fragments, which can be obtained by paired-end sequencing. By using an AMpure size selection, it is possible to enrich for these short fragments and increase the cost-effectiveness of the technique. In contrast, conventional ChIP and ChIP-exo are designed for single-end sequencing. Furthermore, the protocols used to generate sequencing libraries for conventional ChIP-seq and ChIP-exo select against fragments below 100 bp.
Additional files
-
Supplementary file 1
Protocol for high-resolution X-ChIP-seq. Detailed protocol for performing high-resolution X-ChIP-seq in cell lines.
- https://doi.org/10.7554/eLife.09225.009





