The chemokine CXCL13 in lung cancers associated with environmental polycyclic aromatic hydrocarbons pollution
Figures
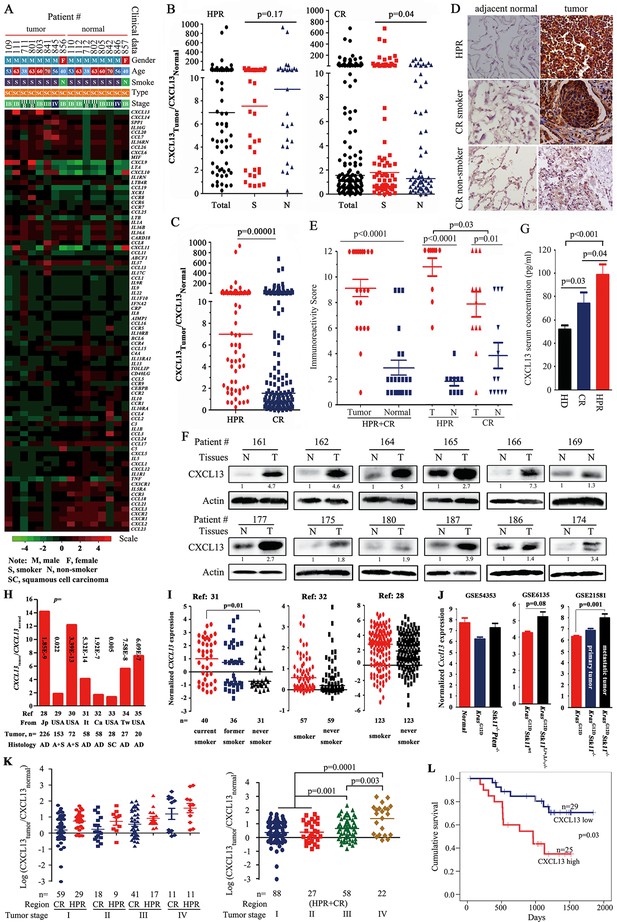
CXCL13 expression in lung cancer.
(A) A PCR array was used to detect the expression of 84 cytokines/chemokines in eight highly polluted region (HPR) lung cancers. (B) The ratios of CXCL13 in tumor samples to their counterpart normal lung tissues from both the HPR and control region (CR) non-small cell lung cancers (NSCLCs). (C) Comparison of the CXCL13tumor/CXCL13normal values of the HPR patients with the CR cases. (D, E) CXCL13 expression was detected by immunohistochemistry (IHC) in HPR and CR patients (D), and the immunoreactivity score was calculated (E). (F) Western blot analyses of lysates from the tumors and adjacent normal lung tissues harvested from CR NSCLCs. (G) The concentrations of CXCL13 in the blood samples from healthy donors (HDs) and HPR and CR patients were detected by ELISA. (H, I) CXCL13 expression in Oncomine reports. (H) CXCL13 expression was detected by microarrays in tumor samples and normal lung tissues. AD, adenocarcinoma; A+S, adenocarcinoma and squamous cell carcinoma; Ca, Canada; It, Italy; Jp, Japan; SC, squamous cell carcinoma; Tw, Taiwan, China. (I) The expression of CXCL13 was detected in tumor tissues of smokers and non-smokers. (J) In mouse Gene Expression Omnibus (GEO) data sets, the expression of cxcl13 in indicated mice was detected by microarray. (K) The relationship between the CXCL13 expression and the tumor stages of lung cancer patients. (L) Overall survival of 54 CR patients (see Table 3 for their baseline demographic characteristics). The median follow-up was 1087 days (range, 187–1845 days).
-
Figure 1—source data 1
Sequences of primers for real-time PCR and ChIP, and siRNA.
- https://doi.org/10.7554/eLife.09419.004
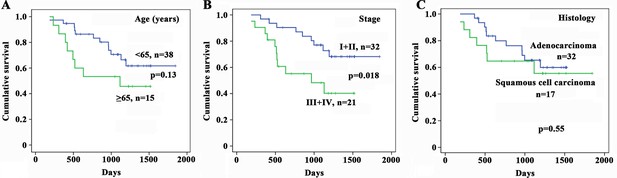
Kaplan–Meier estimates of survival of patients with non-small cell lung cancer (NSCLC) according to age, cancer stage, and histology.
https://doi.org/10.7554/eLife.09419.005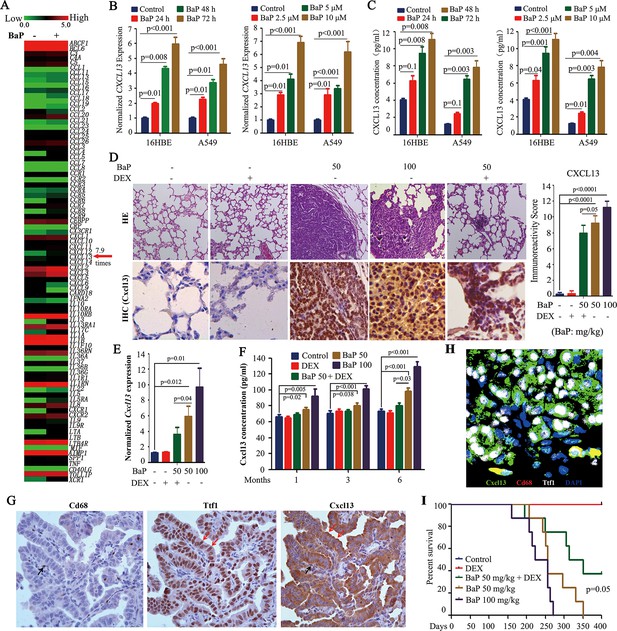
Benzo(a)pyrene (BaP) induces CXCL13 in vitro and in vivo.
(A) A PCR array analysis of the expression of 84 cytokines/chemokines in 16HBE normal lung epithelial cells treated with 1 μM BaP for 30 days. (B) The cells were treated with BaP at 10 μM for indicated time points or with the indicated concentrations for 72 hr, and CXCL13 expression was assessed by real-time RT-PCR. The experiments were conducted in triplicate and repeated three times. The error bars represent the SD. (C) The cells were treated with BaP as described in (B), and the concentration of CXCL13 in the supernatants was evaluated by ELISA. (D) The A/J mice were treated with BaP and/or dexamethasone (DEX) for 5 weeks (see also Figure 2—figure supplement 1A) and sacrificed 6 months later. The lung tissues were isolated and analyzed by Hematoxylin and eosin (HE) staining or immunohistochemistry (IHC) using an anti-Cxcl13 antibody (left panel). The immunoreactivity score was calculated (right panel). (E) Cxcl13 expression was detected in the lung tissues by real-time PCR. (F) The concentration of Cxcl13 in mouse serum was assayed by ELISA. (G) IHC assays of mice’ lung tumor tissues using anti-Cd68, anti-Ttf1, and anti-Cxcl13 antibodies. (H) Immunofluorescence assay of mice’ lung tumor tissues using antibodies against Cxcl13 (green), Cd68 (red), and Ttf1 (white). 4',6-diamidino-2-phenylindole (DAPI) was used to stain the nucleus (blue). (I) The survival curves of the mice treated with BaP and/or DEX (n=8 for each group).
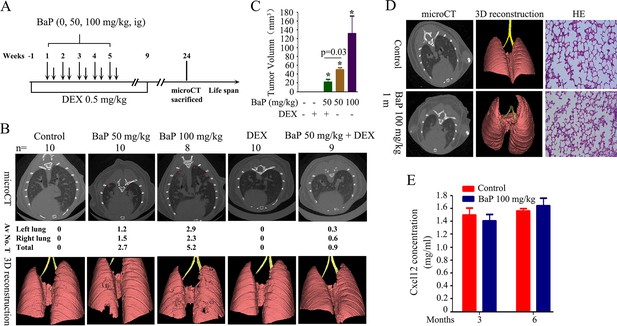
Benzo(a)pyrene (BaP) induces lung cancer in A/J mice.
(A) Schematic represents the protocols for administration of BaP and/or dexamethasone (DEX) in A/J mice. (B) MicroCT and 3D reconstruction of lung cancer in mice treated with BaP and/or DEX. Av No. T, average of numbers of tumors in the mice. (C) Tumor volume of mice treated with BaP and/or DEX. (D) One month after BaP treatment (100 mg/kg), the mice were scanned with microCT and then sacrificed, lung tissues were isolated and analyzed by HE staining. (E) Cxcl12 concentration in peripheral blood of the mice was assayed by ELISA.
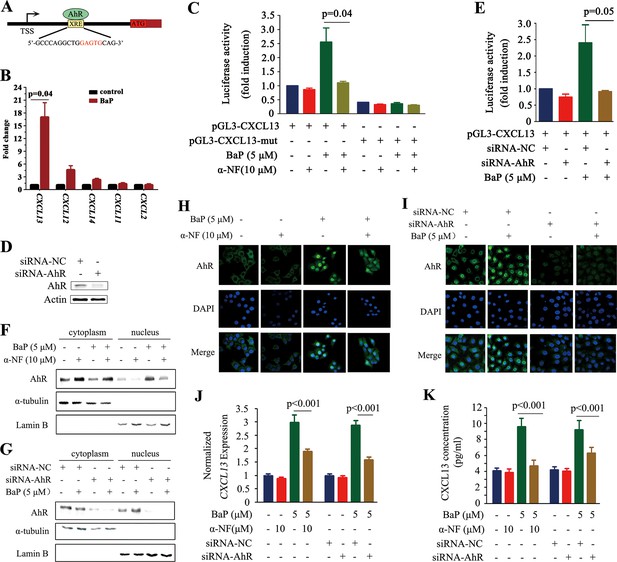
CXCL13 is a target gene of aryl hydrocarbon receptor (AhR).
(A) The AhR binding site is located at 1.7 kb downstream of the CXCL13 transcription start site (TSS). (B) A chromatin immunoprecipitation (ChIP) assay was performed in BaP-treated or untreated 16HBE cells. The enriched CXCL13 was detected by qPCR. (C) The A549 cells were transfected with the wild-type (WT) or mutant (deletion mutation (mut) in the XRE-like sequence) CXCL13 promoter-luciferase reporter construct, treated with BaP and/or α-NF for 48 hr, and assessed by the luciferase assays. (D, E) A549 cells were transfected with AhR-specific siRNAs, and western blot was performed to detect the expression of AhR. Three siRNAs were used, and the result of one was shown (D). Luciferase assays were performed in A549 cells transfected with the WT CXCL13 promoter-luciferase reporter construct and siRNAs in the absence or presence of BaP (E). (F, G) Western blot analyses of AhR in the cytoplasm and nucleus of 16HBE cells co-incubated with BaP, with or without α-NF treatment (F) or siRNA transfections (G). (H, I) Immunofluorescence assays of AhR expression in 16HBE cells co-incubated with BaP, with or without α-NF treatment (H) or siRNA transfections (I). (J, K) CXCL13 mRNA (detected by qPCR; J) and protein (in supernatants of the cells detected by ELISA; K) levels in the AhR-silenced 16HBE cells treated with BaP and/or α-NF.
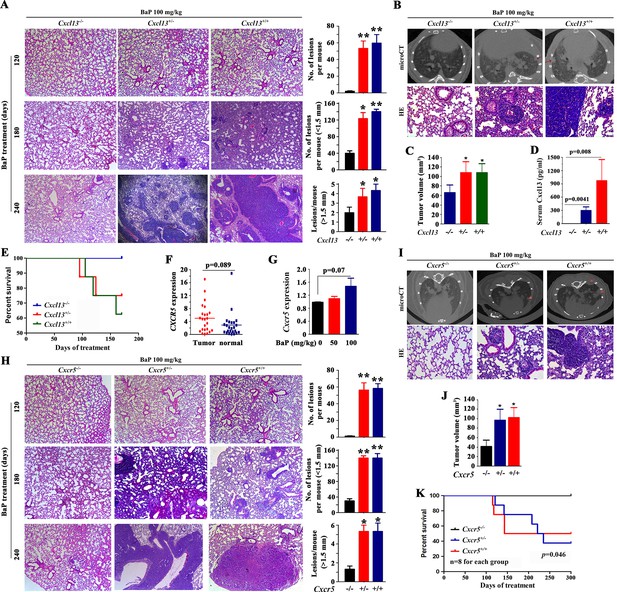
Cxcl13 and Cxcr5 are critical to benzo(a)pyrene (BaP)-induced lung cancer.
(A) Cxcl13 deficiency mice were treated with BaP, sacrificed 120 days, 180 days or 240 days later, and the tumor nodules in histologic sections were analyzed. See also (Figure 4—figure supplement 1). (B) MicroCT scanning images and HE staining of lung sections from the BaP-treated Cxcl13 wild-type (WT) or knockout mice. (C) Tumor volume of the microCT scanning of the mice. (D) Serum concentrations of Cxcl13 in the BaP-treated Cxcl13 WT or knockout mice. (E) Life span of the BaP-treated Cxcl13+/+, Cxcl13+/-, and Cxcl13-/- mice. (F, G) Cxcr5 expression in non-small cell lung cancers (NSCLCs, n=24; F) and in A/J mice treated with BaP (n=6 for each group; G). (H) Cxcr5 deficiency mice were treated with BaP, sacrificed 120 days, 180 days or 240 days later, and the tumor nodules in histologic sections were analyzed. See also Figure 4—figure supplement 1. (I) MicroCT scanning images and HE staining of lung sections from BaP-treated Cxcr5 WT or knockout mice. (J) Tumor volume of the microCT scanning of the mice. (K) Life span of the BaP-treated Cxcr5 WT or knockout mice. *p<0.05; **p<0.01.
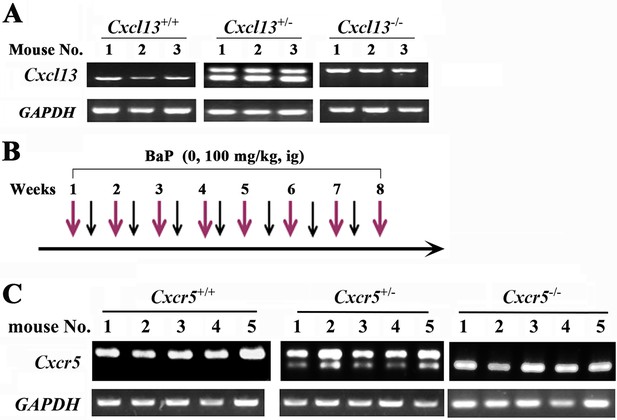
Treatment of Cxcl13-/- and Cxcr5-/- mice with benzo(a)pyrene (BaP).
(A) The expression of Cxcl13 in the mice. (B) Schematic represents the protocols for administration of BaP in the mice. (C) The expression of Cxcr5 in the mice.
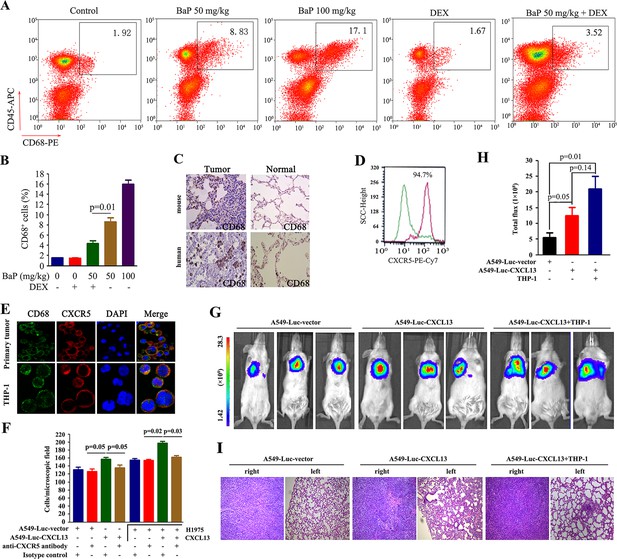
Tumor-associated macrophages in benzo(a)pyrene (BaP)-induced lung cancer.
(A, B) Flow cytometry analysis of Cd68+ macrophages in BaP-induced tumors. A representative gating is shown. The numbers indicate the Cd68+ cells in the quadrant expressed as the percentage of the total Cd45+ leukocytes from the same tumor (A). The means+SD of the Cd68+ cells from the mice (n=10 for each group) are shown (B). See also Figure 5—figure supplement 1. (C) IHC analysis of CD68+ macrophages in tumor samples from BaP-treated mice and highly polluted region (HPR) patients. (D) Flow cytometry analysis of Cd68+ macrophages isolated from tumor samples of mice treated with 50 mg/kg BaP using an anti-Cxcr5 antibody. (E) Immunofluorescence analysis of tumor-associated macrophages in tumor samples from HPR patients and THP-1 cells using anti-CD68 and anti-CXCR5 antibodies; DAPI was used to counterstain the nucleus. (F) A trans-well migration assay was performed by plating THP-1 cells in the lower chambers, and the indicated cells in the upper chambers, with or without anti-CXCR5 antibody. (G, H) Bioluminescent assays of mice that were inoculated with A549- Luciferase (Luc) or A549-Luc-CXCL13 cells (8×105) in the right lung. THP-1 cells (8×105) were injected via the tail vein. Representative images (G) and total luminous flux (H) were shown. (I) Lung sections of the mice were stained with HE.
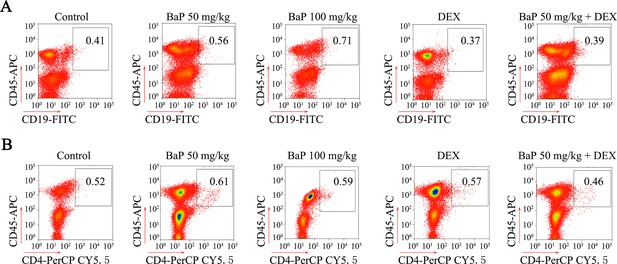
Analysis of B cells and T cells in benzo(a)pyrene (BaP)-induced lung cancer.
(A) The expression of Cd19 in Cd45+ leukocytes sorted from lung cancer tumor samples was analyzed by flow cytometry. (B) The expression of Cd4 was analyzed in Cd45+ leukocytes by flow cytometry.
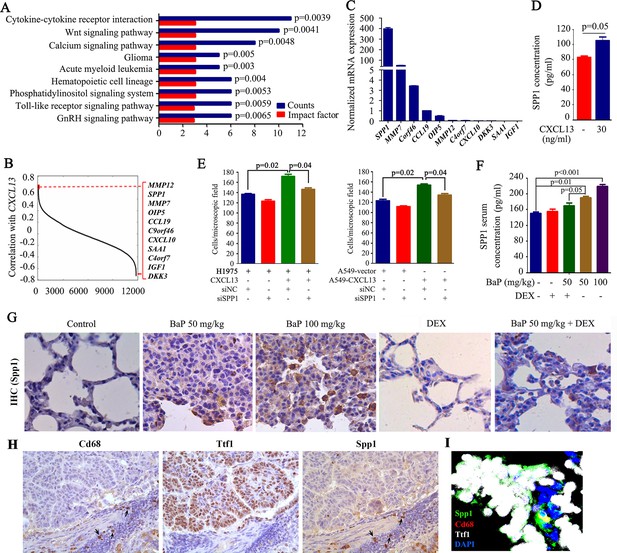
Identification of macrophage-secreted SPP1 as a downstream effector of CXCL13.
(A) The pathway analysis of CXCL13 associated genes. The data from the microarray data sets of the eight highly polluted region (HPR) lung cancers are shown. See also Figure 6—source data 1. (B) Gene ranking according to the correlation with CXCL13 expression. The genes were filtered based on extracellular localization to identify paracrine mediators. The list on the right shows the genes that correlate most significantly with CXCL13. (C) The mRNA expression of the candidate gene was detected in THP-1 cells by qPCR. (D) The concentration of SPP1 in supernatants of THP-1 cells in the absence or presence of CXCL13. (E) Trans-well migration assays were performed by plating THP-1 cells (transfected with siSPP1 or siNC) in the lower chambers, and the indicated cells (CXCL13-treated or untreated) in the upper chambers. (F) Spp1 serum concentrations of mice treated with benzo(a)pyrene (BaP) and/or dexamethasone (DEX) were detected by ELISA. (G) Spp1 expression in lung section of mice treated with BaP and/or DEX was determined by IHC. (H) IHC assays using antibodies against Cd68, Ttf1, and Spp1. (I) Immunofluorescence assay using antibodies against Spp1 (green), Cd68 (red), and Ttf1 (white). DAPI was used to stain the nucleus (blue).
-
Figure 6—source data 1
CXCL13-associated genes in lung cancer.
- https://doi.org/10.7554/eLife.09419.017
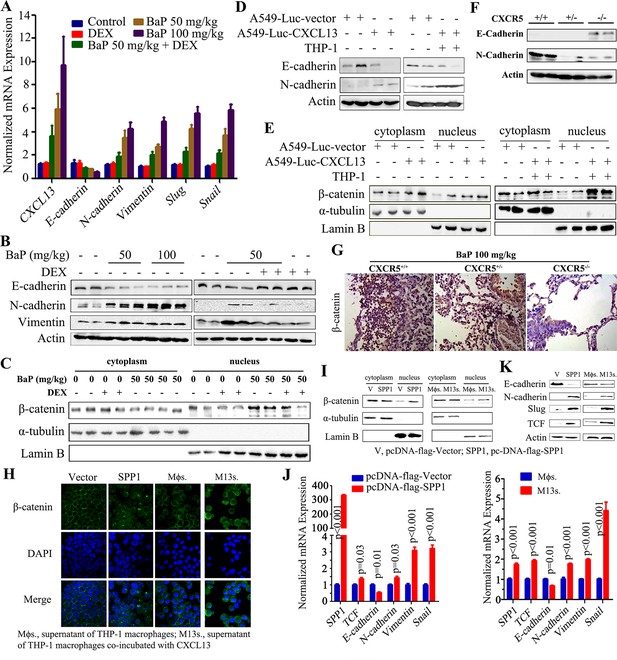
Activation of β-catenin and epithelial mesenchymal transition (EMT) in benzo(a)pyrene (BaP)-induced lung cancer.
(A) The expression of indicated genes in lung tissues from A/J mice treated with BaP and/or dexamethasone (DEX) was detected by real-time PCR. (B) Western blot analyses of lysates of lung tissues from A/J mice treated with BaP and/or DEX. (C) Western blot analyses of cytoplasmic and nucleic protein fractions of lung tissues from A/J mice treated with BaP and/or DEX, using indicated antibodies. (D) Western blot analysis of lysates of lung tissues from NOD/SCID mice injected with indicated cells. (E) Western blot analyses of cytoplasmic and nucleic protein fractions in lung tissues from NOD/SCID mice injected with indicated cells. (F) Western blot analyses of lysates of lung tissues from Cxcr5 mutant mice treated with BaP. (G) IHC assays for the expression of β–catenin in Cxcr5 mutant mice upon BaP. (H) Immunofluorescence analyses of β-catenin in A549 cells transfected with control vector (V) or SPP1, or treated with THP-1 supernatant (Mϕs.) or supernatant of THP-1 cells co-incubated with CXCL13 (M13s.). (I) Western blot analyses of cytoplasmic and nucleic protein fractions of A549 cells transfected with control vector (V) or SPP1, or treated with Mϕs. or M13s. (J) Real-time PCR assays of indicated genes in A549 cells transfected with control vector or SPP1, or treated with Mϕs. or M13s. (K) Western blot analyses of lysates of A549 cells transfected with control vector (V) or SPP1, or treated with Mϕs. or M13s, using indicated antibodies.
Tables
Baseline demographic characteristics of the 201 patients who underwent CXCL13 analyses.
| Characteristics | Total | Highly polluted region (HPR) | Control region (CR) | p values (HPR vs CR) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case, n | CXCL13 high, n (%) | p values | Case, n | CXCL13 high, n (%) | p values | Case, n | CXCL13 high, n (%) | p values | ||
| Total | 201 | 134 (66.7) | 70 | 63 (90) | 131 | 71 (54.2) | 0.0000003 | |||
| G: Male | 134 | 88 (65.7) | 0.67 | 47 | 41 (87.2) | 0.27 | 87 | 47 (54) | 0.95 | 0.0001 |
| Female | 67 | 46 (68.7) | 23 | 22 (95.7) | 44 | 24 (54.5) | 0.0006 | |||
| A: <65 y | 140 | 98 (70) | 0.21 | 56 | 51 (91.1) | 0.55 | 84 | 47 (56) | 0.7 | 0.000009 |
| ≥65 y | 56 | 34 (60.7) | 14 | 12 (85.7) | 42 | 22 (52.4) | 0.03 | |||
| Unknown | 5 | 2 (40) | 5 | 2 (40) | ||||||
| S: Smoker | 107 | 75 (70.1) | 0.18 | 36 | 31 (86.1) | 0.17 | 71 | 44 (62) | 0.04 | 0.01 |
| Non-smoker | 87 | 53 (60.9) | 27 | 26 (96.3) | 60 | 27 (45) | 0.000006 | |||
| Unknown | 7 | 6 (85.7) | 7 | 6 (85.7) | ||||||
| H: Adenocarcinoma (AD) | 131 | 91 (69.5) | 0.45 | 48 | 44 (91.7) | 0.37 | 83 | 47 (56.6) | 0.84 | 0.00003 |
| Squamous cell carcinoma (SCC) | 61 | 39 (63.9) | 19 | 16 (84.2) | 42 | 23 (54.8) | 0.03 | |||
| Others | 9 | 4 (44.4) | 3 | 3 (100) | 6 | 1 (16.7) | ||||
| Tumor node metastasis (TNM): I | 88 | 51 (58) | 0.007 | 29 | 23 (79.3) | 0.02 | 59 | 28 (47.5) | 0.05 | 0.004 |
| II | 27 | 17 (63) | 9 | 8 (88.9) | 18 | 9 (50) | 0.005 | |||
| III | 58 | 43 (74.1) | 17 | 17 (100) | 41 | 26 (63.4) | 0.004 | |||
| IV | 22 | 19 (86.4) | 11 | 11 (100) | 11 | 8 (72.7) | 0.06 | |||
| Unknown | 6 | 4 (66.7) | 4 | 4 (100) | 2 | 0 (0) | ||||
-
G, gender; A, age; S, smoke; H, histology.
Multivariate logistic analyses of the association between CXCL13 high expression and clinical characteristics.
| Highly polluted region (HPR) patients, n=70 | |||
| Variable | Odds ratio | 95.0% confidence interval | P value |
| Age | 1.483 | 0.177–12.452 | 0.716 |
| Gender | 4.711 | 0.488–45.51 | 0.18 |
| Smoking | 0.652 | 0.115–3.682 | 0.628 |
| Histology | 0.37 | 0.069–1.992 | 0.247 |
| TNM stage | 3.092 | 0.765–12.502 | 0.113 |
| Control region (CR) patients, n=131 | |||
| Variable | Odds ratio | 95.0% confidence interval | P value |
| Age | 0.914 | 0.455–1.832 | 0.799 |
| Gender | 2.15 | 0.772–5.993 | 0.143 |
| Smoking | 0.513 | 0.251–1.052 | 0.06 |
| Histology | 1.148 | 0.569–2.313 | 0.7 |
| TNM stage | 2.355 | 1.157–4.793 | 0.018 |
| Total (HPR and CR patients), n=201 | |||
| Variable | Odds ratio | 95.0% confidence interval | P value |
| Region | 7.908 | 3.272–19.114 | 4.6×10-6 |
| Age | 0.964 | 0.5–1.861 | 0.914 |
| Gender | 2.254 | 0.897–5.662 | 0.084 |
| Smoking | 0.394 | 0.168–0.925 | 0.032 |
| Histology | 0.996 | 0.52–1.907 | 0.99 |
| TNM stage | 2.707 | 1.401–5.232 | 0.003 |
Baseline demographic characteristics of 54 control region (CR) lung cancer patients whose survival information was available.
| Characteristics | Case, n | CXCL13-high, n (%) | P* Value |
|---|---|---|---|
| Total | 54 | 25 (46.3) | |
| Gender | |||
| Male | 38 | 16 (42.1) | 0.34 |
| Female | 16 | 9 (56.3) | |
| Smoking | |||
| Smoker | 33 | 16 (48.5) | 0.69 |
| Non-smoker | 21 | 9 (42.9) | |
| Age, years | 0.72 | ||
| <65 | 38 | 17 (44.7) | |
| ≥65 | 16 | 8 (50) | |
| Histology | |||
| Adenocarcinoma | 32 | 15 (46.9) | 0.83 |
| squamous cell carcinoma | 18 | 9 (50) | |
| small cell lung cancer | 1 | 0 (0) | |
| Others | 3 | 1 (33.3) | |
| TNM stage | 0.65 | ||
| I | 26 | 12 (46.2) | |
| II | 6 | 2 (33.3) | |
| III | 18 | 9 (50) | |
| IV | 4 | 2 (50) |





