Precise let-7 expression levels balance organ regeneration against tumor suppression
Figures
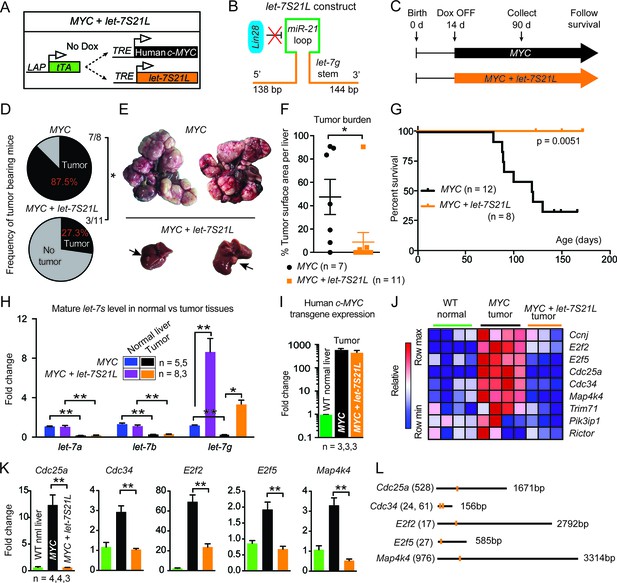
let-7g inhibits the development of MYC-driven hepatoblastoma.
(A) Schema of the liver-specific inducible LAP-MYC +/- let-7S21L cancer model. (B) let-7S21L is a chimeric construct containing the let-7g stem, miR-21 loop, and let-7g flanking sequences. (C) Schema showing that LAP-MYC +/- let-7S21L mice were induced at 14 days of age, tissues were collected at 90 days of age, and survival was followed. (D) Ninety-day old mice bearing tumors in the LAP-MYC (87.5%, 7/8) and LAP-MYC + let-7S21L (27.3%, 3/11) mouse models. (E) Livers showing tumors from the above mice. (F) Liver surface area occupied by tumor. (G) Kaplan-Meier curve of LAP-MYC alone and LAP-MYC + let-7S21L mice. (H) Mature let-7 expression levels in as determined by RT-qPCR. (I) Human c-MYC mRNA expressionin tumors as determined by RT-qPCR. (J) Heat map of let-7 target gene expression in WT normal livers, MYC tumors, and MYC + let-7S21L tumors as measured by RT-qPCR. Red is higher and blue is lower relative mRNA expression. (K) Gene expression plotted as bar graphs to show relative changes. (L) Evolutionarily conserved let-7 target sites within 3’UTRs (Targetscan.org). All data in this figure are represented as mean ± SEM. *p < 0.05, **p < 0.01.
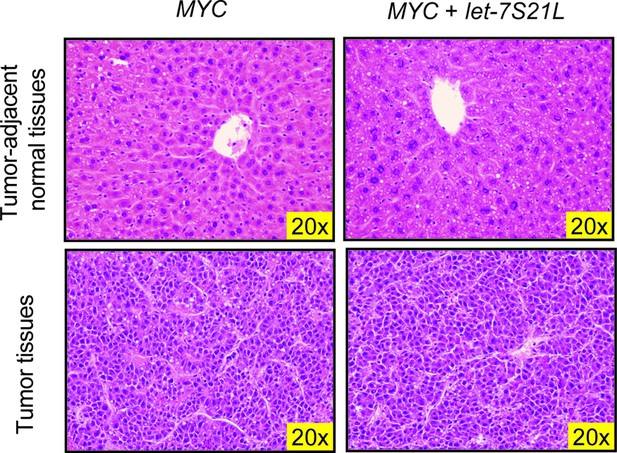
H&E staining of LAP-MYC and LAP-MYC + let-7S21L tumor-adjacent normal tissues and tumor tissues.
https://doi.org/10.7554/eLife.09431.004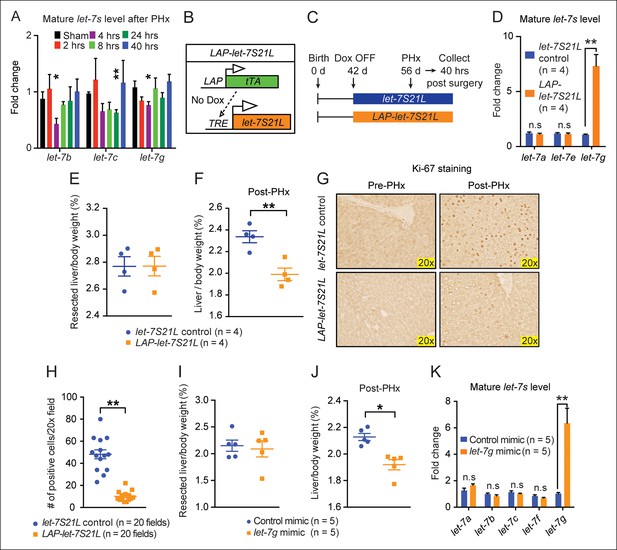
let-7g overexpression inhibits liver regeneration after partial hepatectomy.
(A) Mature endogenous let-7 expression levels in WT C57Bl/6 mice at different time points after PHx as determined by RT-qPCR (n=4 and 4 for each time point). (B) Schema of the LAP-let-7S21L dox-inducible model. LAP-tTA single transgenic mice served as the controls. (C) Schema showing that let-7S21L control and LAP-let-7S21L mice were induced at 42 days of age, PHx was performed after 14 days of induction, and tissues were collected 40 hr post PHx. (D) Mature let-7 expression levels in let-7S21L and LAP-let-7S21L livers after 14 days of induction (n=4 and 4). (E) Resected liver/body weight ratios of LAP-tTA Control and LAP-let-7S21L mice at the time of PHx (n=4 and 4). (F) Liver/body weight ratios 40 hr after PHx (n=4 and 4). (G) Ki-67 staining on resected and post-PHx liver tissues. (H) Quantification of Ki-67-positive cells (n=2 and 2 mice; ten 20x fields for each mouse were quantified). (I) Resected liver/body weight ratios 2 days after intravenous injection of 0.5 mg/kg negative control or let-7g microRNA mimics packaged in C12-200 LNPs (n=5 and 5). (J) Liver/body weight ratios 40 hr after PHx (n=4 and 4). (K) Mature let-7g expression levels in mimic treated livers (n=5 and 5). All data in this figure are represented as mean ± SEM. *p<0.05, **p<0.01.
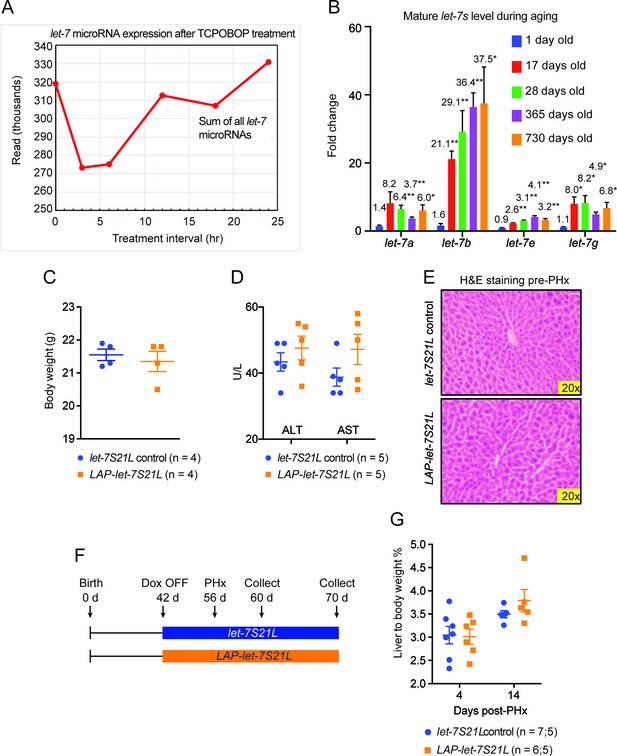
Data associated with Figure 2.
(A) Sum of the absolute sequencing reads for mature let-7 microRNA family members after TCPOBOP treatment, as measured by small RNA sequencing. (B) Mature let-7 microRNA family expression in WT mouse livers at different ages as determined by RT-qPCR. Numbers over bars indicate the fold change normalized to that of 1 day old mice. (C) Body weights of let-7S21L alone and LAP-let-7S21L mice pre-PHx (n=4 and 4). (D) Liver function tests: ALT (U/L) and AST (U/L) of let-7S21L alone and LAP-let-7S21L mice pre-PHx (n=5 and 5). (E) H&E staining of let-7S21L alone and LAP-let-7S21L mice pre-PHx. (F) Schema showing that let-7S21L control and LAP-let-7S21L mice were induced at 42 days of age, PHx was performed after 14 days of induction, and tissues were collected 4 and 14 days after PHx. (G) Liver to body weight ratio of let-7S21L alone and LAP-let-7S21L mice 4 and 14 days after PHx (n=5 and 5). All data in this figure are represented as mean ± SEM. *p<0.05, **p<0.01.
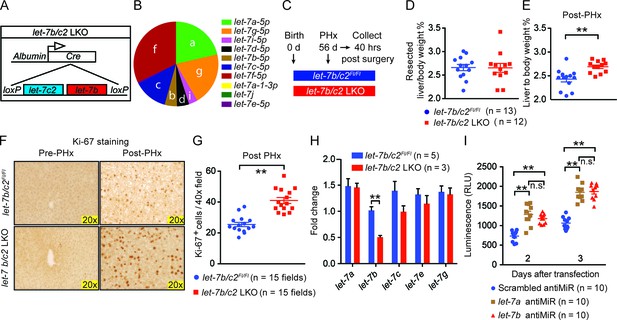
Loss of let-7b and let-7c2 is sufficient to enhance liver regeneration.
(A) Schema of liver-specific let-7b and let-7c2 knockout mice (let-7b/c2 LKO). Albumin-Cre excises loxPs in the embryonic liver of let-7b/c2Fl/Fl mice. Mice without Cre serve as the controls. (B) Small RNA sequencing showing the distribution of 10 let-7s in WT mice (n=2) (Data obtained from Xie et al. 2012). (C) Schema showing that PHx was performed on let-7b/c2Fl/Fl and let-7b/c2 LKO mice at 56 days of age and tissues were collected 40 hr post PHx. (D) Resected liver/body weight ratios at the time of PHx, and (E) Liver to body ratios of let-7b/c2Fl/Fl (n=11) and let-7b/c2 LKO mice (n=10) 40 hr after PHx. (F) Ki-67 staining and (G) Quantification of Ki-67-positive cells on resected and 40 hr post-PHx liver tissues (n=3 and 3 mice; total of five 40x fields/mouse were used for quantification). (H) RT-qPCR on let-7 family members from let-7b/c2Fl/Fl and let-7b/c2 LKO mice pre- and 40 hr post-PHx. (I) Viability of H2.35 immortalized human hepatocytes treated with either scrambled, let-7a, or let-7b antiMiRs, measured at two and three days after transfection (n=10 each). All data in this figure are represented as mean ± SEM. *p<0.05, **p<0.01.
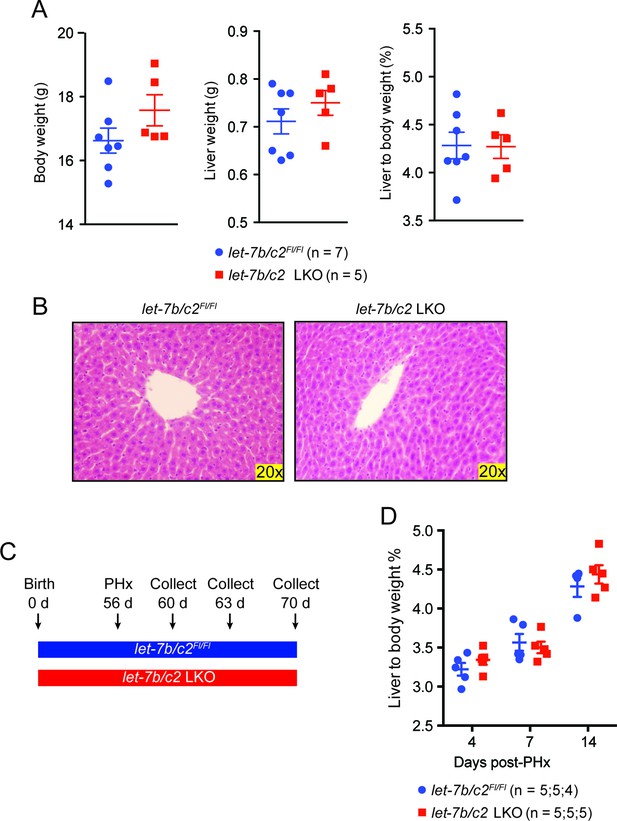
Characterization of let-7b/c2 LKO mice.
(A) Body weight, liver weight, and liver to body weight percent of let-7b/c2Fl/Fl (n = 7) and let-7b/c2 LKO mice (n=5) at 56 days of age. (B) H&E staining of let-7b/c2Fl/Fl (n=3) and let-7b/c2 LKO livers (n=3). (C) Schema showing that PHx was performed on let-7b/c2Fl/Fl and let-7b/c2 LKO mice at 56 days of age and tissues were collected at 4, 7, and 14 days post PHx. (D) Liver to body weight percent of let-7b/c2Fl/Fl (n=11) and let-7b/c2 LKO mice (n=10) 4, 7, and 14 days after PHx. All data in this figure are represented as mean ± SEM. *p<0.05, **p<0.01.
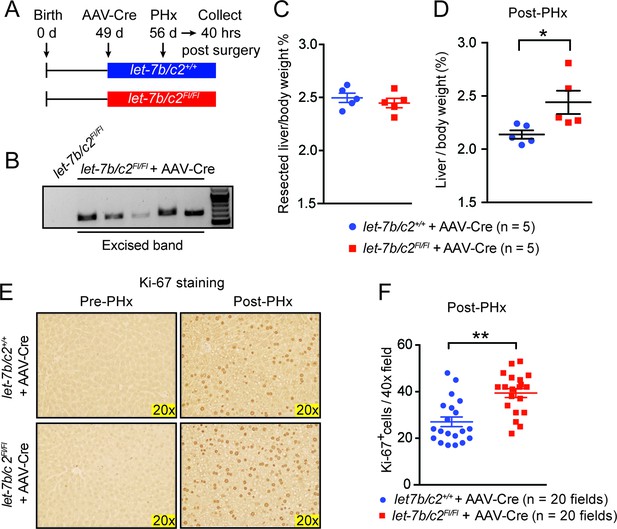
Post-natal deletion of let-7b/c2 also enhances liver regeneration.
(A) Schema showing that let-7b/c2 +/+ and let-7b/c2Fl/Fl mice were injected with AAV-Cre at 49 days of age, PHx was performed 7 days after viral injection, and tissues were collected 40 hr post PHx. (B) DNA gel showing excised let-7b/c2 band in let-7b/c2Fl/Fl + AAV-Cre mice (n=5) but not in let-7b/c2 +/+ + AAV-Cre mice. (C) Percentage of resected liver/body weight ratios of let-7b/c2 +/+ + AAV-Cre (n=5) and let-7b/c2Fl/Fl + AAV-Cre mice (n=5) at the time of PHx. (D) Liver/body weight ratios of the above mice 40 hr after PHx. (E) Ki-67 staining on resected and 40 hr post-PHx livers from the above mice (n=3 and 3). (F) Quantification of Ki-67-positive cells (n=2 and 2 mice; total of ten 40x fields/mouse were used for quantification). All data in this figure are represented as mean ± SEM. *p<0.05, **p<0.01.
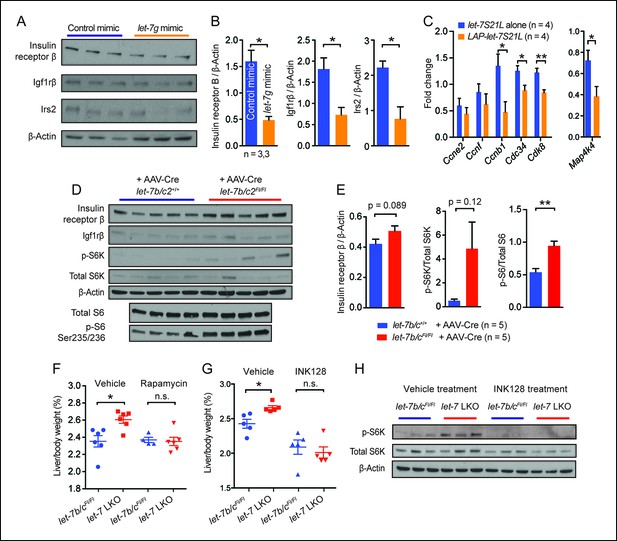
let-7g suppresses liver regeneration through insulin-PI3K-mTOR.
(A) Western blots of insulin receptor β, Igf1rβ, Irs2, and β-Actin in negative control or let-7g microRNA mimic treated liver tissues 40 hr after PHx. (B) Quantification of intensity of insulin receptor β, Igf1rβ, Irs2 (Image J). (C) Cell cycle gene expression in let-7S21L alone (n=4) and LAP-let-7S21L (n=4) livers before and 40 hr after PHx as determined by RT-qPCR. (D) Western blots of insulin receptor β, Igf1rβ, p-S6K, total S6K, β-Actin, p-S6 (Ser235/236), and total S6 in AAV-Cre treated let-7b/c2 +/+ and let-7b/c2Fl/Fl livers (n=5 and 5). (E) Quantification of intensity of insulin receptor β/β-Actin, p-S6K/total S6K, and p-S6/total S6, 40 hr after PHx (Image J). (F) Rapamycin treatment during and after PHx in let-7b/c2Fl/Fl control and let-7b/c2 LKO mice. Shown are liver weights 40 hr post PHx. (G) INK128 treatment during and after PHx in let-7b/c2Fl/Fl control and let-7b/c2 LKO mice. Shown are liver weights 40 hr post PHx. (H) Western blots of p-S6K, total S6K, and β-Actin in let-7b/c2Fl/Fl control and let-7b/c2 LKO livers treated with either vehicle or INK128 at 40 hr post PHx. All data in this figure are represented as mean ± SEM. *p<0.05, **p<0.01.
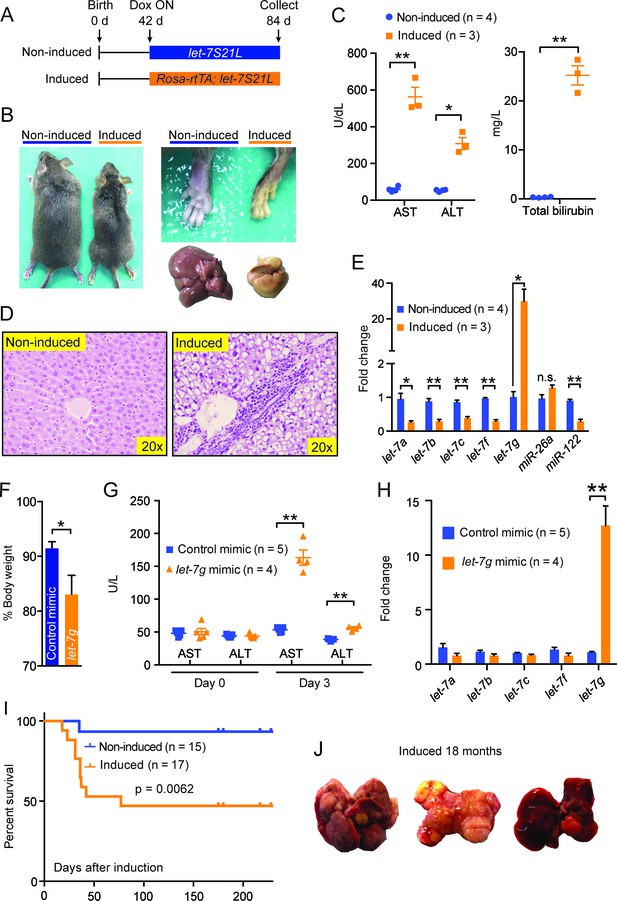
Chronic high-dose let-7g causes hepatotoxicity and liver carcinogenesis.
(A) Schema showing that let-7S21L control and Rosa-rtTA; let-7S21L mice were induced at 42 days of age and collected at 84 days. (B) Images showing the whole body, extremities, and livers of Rosa-rtTA (n=4) and Rosa-let-7S21L mice (n=3) given 1 mg/mL dox between 42 and 84 days of age. (C) Liver function tests: AST (U/L), ALT (U/L), and total bilirubin (mg/L) in these mice. (D) H&E staining of livers. (E) RT-qPCR of mature let-7s and other microRNAs in let-7g overexpressing mice (n=4 and 3). (F) Body weight 3 days after injection of 2.0 mg/kg negative control or let-7g microRNA mimics packaged in C12-200 LNPs relative to pre-injection weight (n=5 and 4). (G) Liver function tests: AST (U/L) and ALT (U/L) in WT C57Bl/6 mice before and 3 days after mimic injection (n=5 and 4). (H) Mature let-7 levels in wild-type C57Bl/6 mice treated with mimics as determined by RT-qPCR (n=5 and 4). (I) Kaplan-Meier curve for Rosa-let-7S21L induced with 1.0 g/L dox at 6 weeks old (n=15 and 17). (J) Gross images of the liver of Rosa-let-7S21L mice induced for 18 months. All data in this figure are represented as mean ± SEM. *p<0.05, **p<0.01.
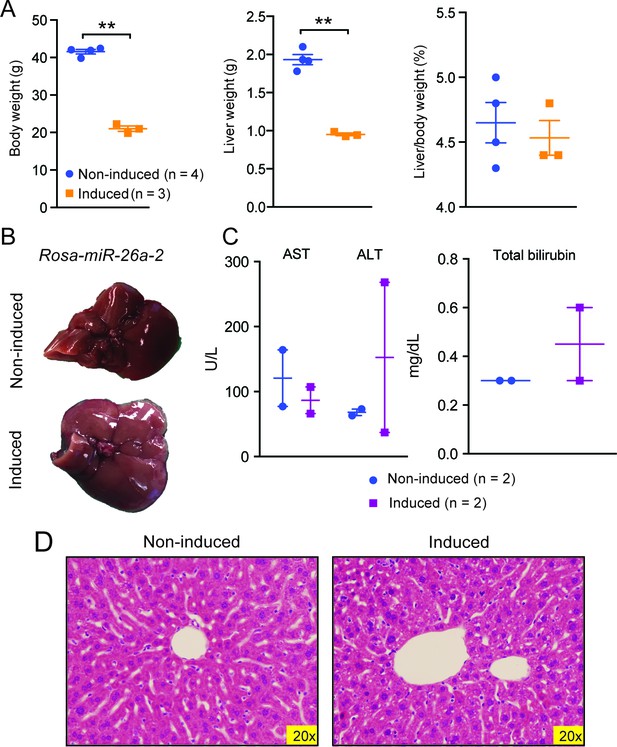
Data associated with Figure 5.
(A) Body, liver, and liver/body weight ratios of control (n=4) and Rosa-let-7S21L (n=3) mice after 42 days of induction. (B) Gross image of Rosa-rtTA (non-induced) and Rosa-miR26a-2 (induced) mice under 1.0 g/L dox for 42 days. (C) Liver function tests: AST (U/L), ALT (U/L), and total bilirubin (mg/L) (n=2 and 2) after induction. (D) H&E staining of Rosa-rtTA (non-induced) and Rosa-miR26a-2 (induced) mice after induction. All data in this figure are represented as mean ± SEM. *p<0.05, **p<0.01.





