Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis
Figures
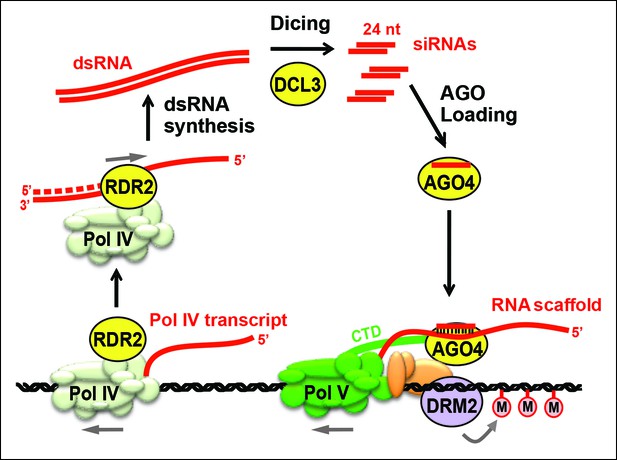
Biogenesis of 24 nt siRNAs and their role in RNA-directed DNA methylation.
A simplified cartoon of the RNA-directed DNA methylation pathway. Polymerase (Pol) IV and RNA-dependent RNA polymerase (RDR2) physically associate and are required for the synthesis of double-stranded RNAs (dsRNA) that are diced by DICER-like 3 (DCL3) into 24 nt siRNA duplexes. Upon loading into Argonaute 4 (AGO4), the siRNA-AGO4 complex finds its target sites by binding to Pol V transcripts and by interacting with the C-terminal domain (CTD) of the Pol V largest subunit. The cytosine methyltransferase DRM2 is ultimately recruited to Pol V-transcribed loci, resulting in de novo cytosine methylation in all sequence contexts (CG, CHG and CHH; where H represents a nucleotide other than G).
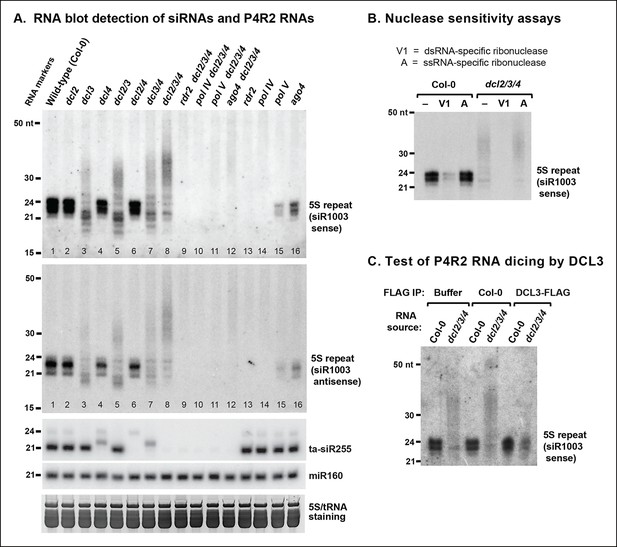
RNA blot analyses of 24 nt siRNAs and their precursors.
(A) The small RNA blot was successively hybridized to probes representing either strand of the siR1003 duplex, a small interfering RNA (siRNA) that is derived from intergenic regions separating 5S ribosomal RNA (rRNA) gene repeats (top two images), as well as to a trans-acting siRNA (ta-siR255) and a microRNA (miR160) probe. An image of the stained gel under fluorescent illumination (in the region that includes 5S rRNA and transfer RNAs [tRNAs]) is shown at the bottom as a loading control. (B) RNA blot of small RNAs isolated from wild-type (ecotype Col-0) or dcl2 dcl3 dcl4 triple mutant (dcl2/3/4) plants, with or without prior treatment with ribonuclease V1 or ribonuclease A. The blot was hybridized to a probe designed to detect siR1003 ‘sense’, which arises from 5S rRNA gene intergenic spacers. (C) Dicing of precursor RNAs by DICER-like 3 (DCL3) in vitro. RNA isolated from wild-type (ecotype Col-0) or from dcl2/3/4 triple mutant plants was incubated with anti-FLAG resin that had been incubated with protein extraction buffer, a cell-free extract of wild-type (Col-0) plants, or a cell-free extract of transgenic plants expressing FLAG-tagged DCL3. RNAs were then purified, subjected to blotting and hybridized to the siR1003 ‘sense’ probe.
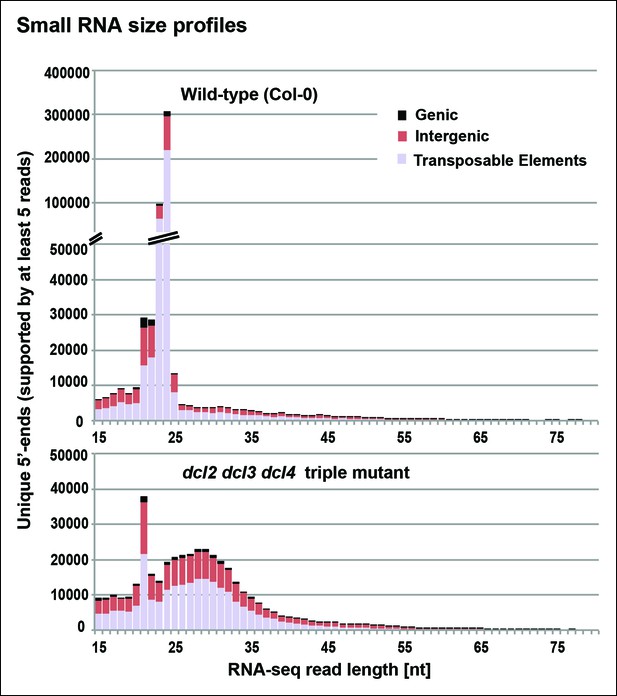
Small RNA size profiles in wild-type and dcl2 dcl3 dcl4 (dcl2/3/4) triple mutants.
The number of unique sequences starting at a given 5’ terminal nucleotide (and represented by at least five reads after normalization and filtering) is plotted for RNAs in each size class. Color coding in each bar depicts the relative proportions of RNA sequencing (RNA-seq) reads that correspond to genes, intergenic regions, or transposable elements. Figure 3—figure supplement 1 provides related data, showing the total numbers of unique 15–94 nt RNA sequences (one or more copies) detected in wild-type or dcl2/3/4 mutants.
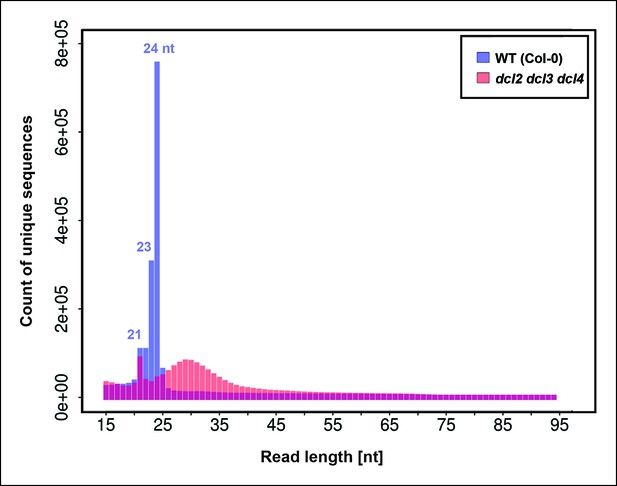
Number of unique sequences among RNAs of 15–94 nt in wild-type or dcl2 dcl3 dcl4 (dcl2/3/4/) triple mutants.
RNAs representing a specific sequence and a particular strand polarity were counted only once, regardless of the actual abundance of RNA sequencing (RNA-seq) reads for that sequence in the dataset. RNA-seq counts for wild-type are shown in blue. RNA-seq counts for dcl2 dcl3 dcl4 mutants are shown in orange. Overlapping read counts are shown in deep red.
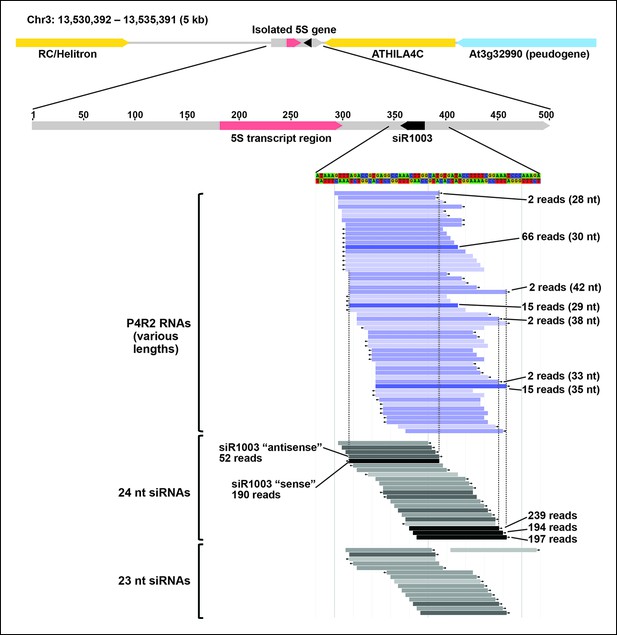
Browser view of Pol IV/RDR2-dependent RNAs (P4R2 RNAs) and 24/23 nt small interfering RNA (siRNAs) in the intergenic spacer region of a 5S ribosomal RNA (rRNA) gene repeat unit.
An isolated 5S rRNA gene repeat (∼500 bp, gray horizontal bar with red transcript region) is shown within its 5 kb chromosomal context, flanked by two transposable elements, shown in yellow, and a pseudogene, shown in blue. Below the diagram, P4R2 RNAs are depicted as horizontal bars shown in shades of blue whereas 24 and 23 nt siRNAs are shown in shades of gray to black, with color intensity reflecting abundance (read counts are provided for several examples). Each bar represents a specific RNA sequence, with arrows depicting the RNA strand orientation relative to the reference genome sequence (TAIR10). Dotted vertical lines provide alignments and show that the ends of highly abundant (>100 reads) siRNA species tend to coincide with the ends of P4R2 RNAs for which there is more than a single read.
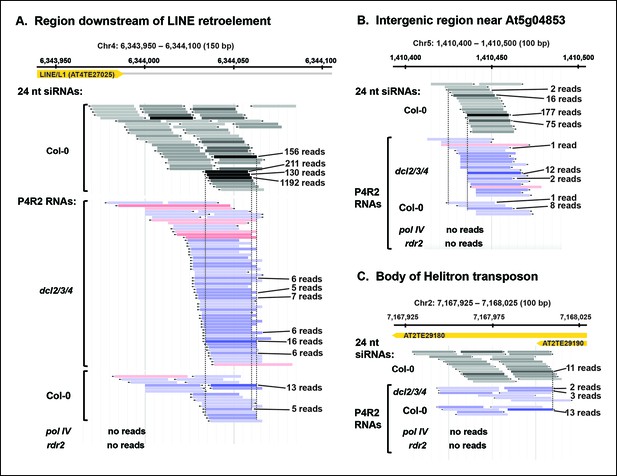
Pol IV/RDR2-dependent RNAs (P4R2 RNAs) are dependent on both Pol IV and RDR2.
(A–C) Browser views of three 24 nt small interfering RNA (siRNA) loci at which abundant P4R2 RNAs that accumulate in dcl2 dcl3 dcl4 (dcl2/3/4) mutants are also observed in wild-type plants (Col-0) but not in nrpd1 (Pol IV) or rdr2 mutants. These examples are representative of a subset of loci selected by virtue of having five or more reads for at least one of the P4R2 species at the locus in wild-type (Col-0) plants. These loci tend to correspond to loci giving rise to abundant siRNAs. Vertical dotted lines provide alignments between abundant siRNAs and the P4R2 RNAs. P4R2 RNAs that are 40 nt or longer are shown in shades of pink. RNAs of 25-39 nt are shown in shades of blue. Browser views for three additional representative loci are shown in Figure 5—figure supplement 1 . Supplementary file 1 provides a table with coordinates for thousands of 100 bp genomic intervals in which 24 nt siRNA loci and putative P4R2 RNAs are detected in wild-type and dcl2/3/4, but are depleted in nrpd1 and rdr2 mutants.
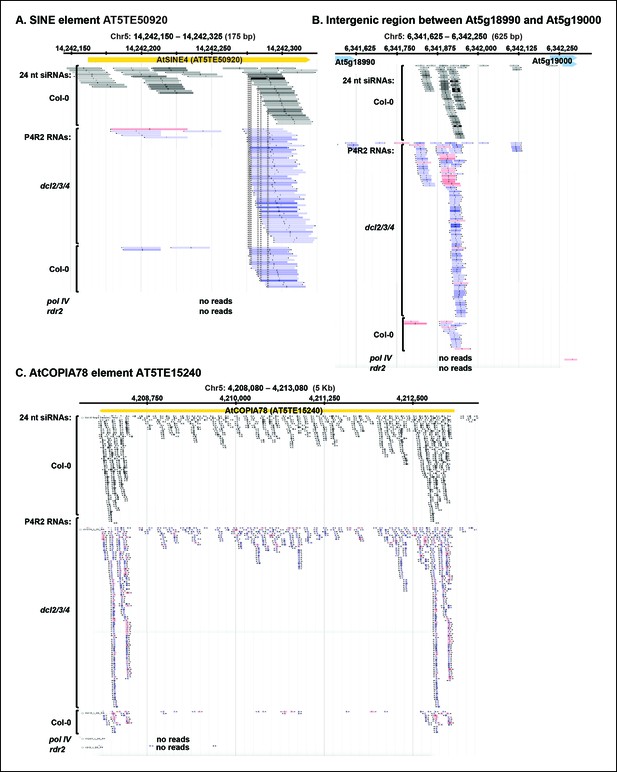
P4R2 RNAs are co-dependent on Pol IV and RDR2.
(A–C) Browser views of three additional 24 nt small interfering RNA (siRNA) loci at which Pol IV/RDR2-dependent RNAs (P4R2 RNAs) that accumulate in dcl2 dcl3 dcl4 (dcl2/3/4) mutants are also observed in wild-type (Col-0) plants (with five or more reads for at least one P4R2 species) but are not observed in nrpd1 polymerase IV (Pol IV) or rdr2 mutants.
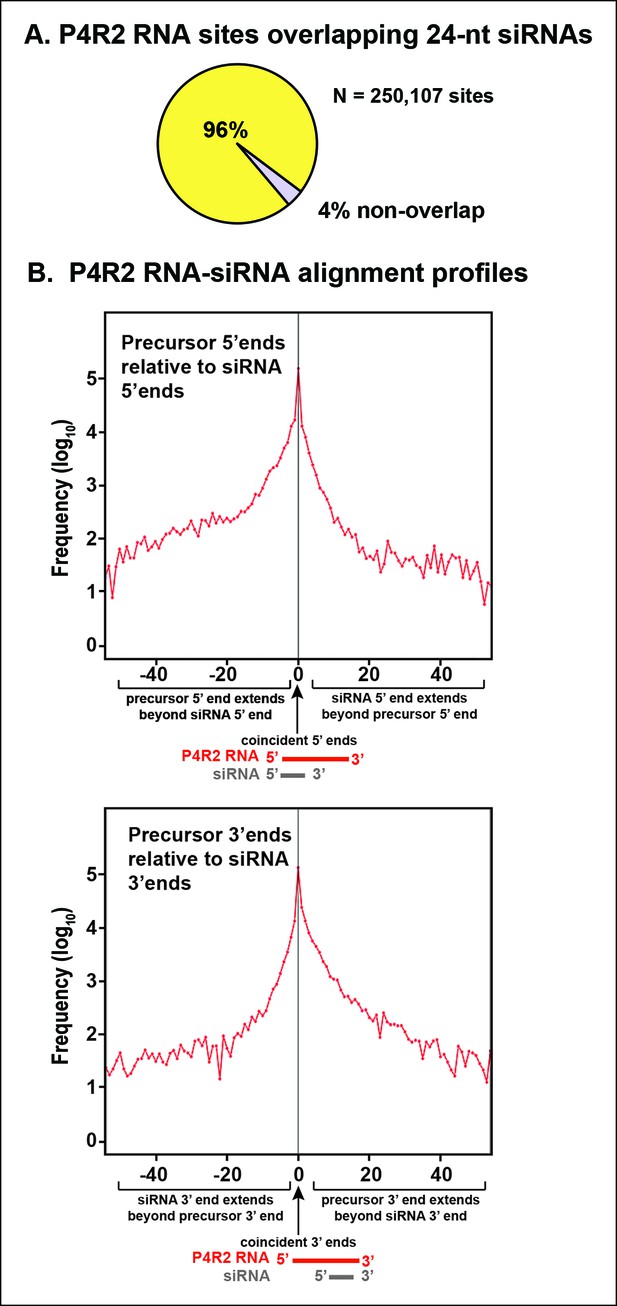
Sequence relationships between Pol IV/RDR2-dependent RNAs (P4R2 RNAs) and small interfering RNA (siRNAs).
(A) Correspondence between P4R2 RNA and siRNA loci. P4R2 RNAs were mapped to the Arabidopsis reference genome (TAIR10) and the frequency at which 24 nt siRNAs overlap these P4R2 genomic positions was calculated. To be considered for this analysis, specific siRNAs had to be represented by at least five reads in wild-type Col-0. Supplementary file 2 shows that P4R2 RNA loci include loci confirmed by Li et al., 2015 to generate Pol IV-dependent transcripts. (B) P4R2 RNA and 24 nt siRNA spatial relationships. The top panel shows the frequency distribution of P4R2 RNA 5’ end positions relative to siRNA 5’ ends. At position zero on the x-axis, P4R2 RNAs and siRNAs share the same 5’ terminus. Negative values indicate how far (in nucleotides) the 5’ end of the P4R2 RNA is located upstream of an siRNA start position. Likewise, positive values indicate how far the 5’ end of a P4R2 RNA is located downstream of an siRNA start position. The lower panel shows the frequency with which P4R2 RNAs and siRNAs align at 3’ ends. At position zero on the x-axis, P4R2 RNAs and siRNAs share the same 3’ terminus. Negative values occur when P4R2 RNAs end upstream of siRNA 3’ ends, and positive values occur when P4R2 RNAs end downstream of siRNA 3’ ends (computed using FEATnotator, v1.2.2, Podicheti and Mockaitis, 2015).
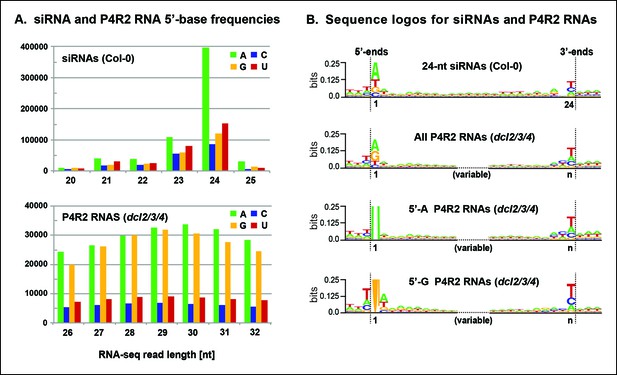
Sequence features of 24 nt small interfering RNAs (siRNAs) and Pol IV/RDR2-dependent RNA (P4R2 RNA) precursors.
(A) Frequencies at which the four nucleotides are present at the 5’ terminus of 20–25 nt small RNAs in wild-type (Col-0) plants or at the 5’ termini of P4R2 RNAs in dcl2 dcl3 dcl4 triple mutants (dcl2/3/4). Data for the subset of P4R2 RNAs in the peak size range of 26–32 nt is shown. (B) Sequence logos for chromosomal DNA sequences corresponding to all 24 nt siRNAs (in wild-type, Col-0), all P4R2 RNAs, P4R2 RNAs that begin with adenosine (5’-A) or P4R2 RNAs that begin with guanosine (5’-G). The P4R2 logos were generated using dcl2/3/4 triple mutant data. The logos include three nucleotides upstream and three nucleotides downstream of the chromosomal DNA sequences that match the RNAs. Each unique RNA sequence is represented only once in the input data. For reads mapping to multiple loci, upstream and downstream DNA sequences were obtained from one mapped site selected at random. Graphics were generated using WebLogo v2.8.2 (Crooks et al., 2004). Figure 7—figure supplement 1 provides sequence logos for P4R2 RNAs of varying length, showing that consensus sequences are consistent among these RNAs.
Sequence logos for Pol IV/RDR2-dependent RNAs (P4R2 RNAs) of different lengths.
P4R2 RNAs of discrete lengths (e.g., 26, 27, or 28 nt, etc.) were analyzed for potential consensus sequences, with results displayed as in Figure 7.
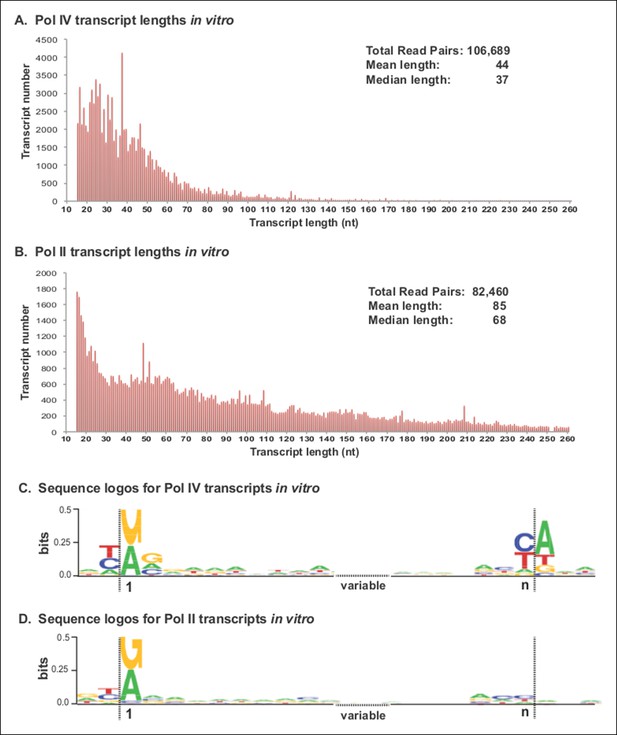
Pol IV transcripts generated in vitro share features of Pol IV/RDR2-dependent RNAs (P4R2 RNAs) in vivo.
(A and B) Size and frequency of RNAs transcribed by polymerase (Pol) IV or II in vitro. Pol IV and Pol II were affinity purified by virtue of FLAG epitope tags fused to the C-termini of the NRPD1 or NRPB2 subunits, respectively. In the case of Pol IV, the transgenic NRPD1-FLAG line is null for the endogenous NRPD1 and RDR2 genes, such that Pol IV is free of associated RDR2. Transcripts generated using closed-circular single-stranded M13 virus as the DNA template were subjected to RNA-seq. The frequency and sizes of mapped reads are plotted. (C and D) Sequence logos for the 5’ and 3’ ends of Pol IV and II in vitro transcripts. RNA-seq, RNA sequencing.
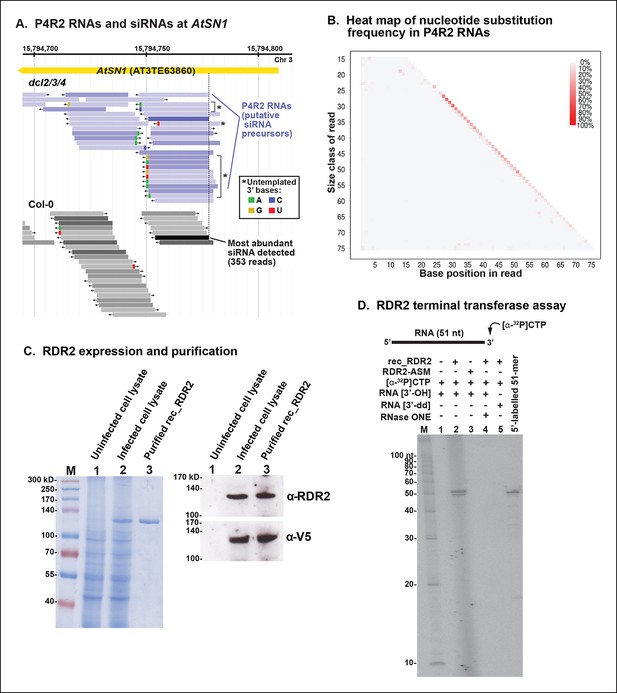
3’ mismatches detected in 24 nt siRNAs and Pol IV/RDR2-dependent RNAs (P4R2 RNAs) may reflect RDR2 terminal transferase activity.
(A) Genome browser view of P4R2 RNAs (shades of blue) and 24 nt small interfering RNA (siRNAs) (shades of gray) at a representative locus, an AtSN1 retrotransposon on chromosome 3. Each horizontal bar represents a specific RNA sequence (RNA-seq), with arrows depicting their direction relative to the Arabidopsis reference genome sequence (TAIR10). The intensity of shading reflects the abundance of each RNA species in the RNA-seq dataset. Brightly colored nucleotides, color coded for A, G, C, or U (see inset), represent nucleotides that do not match the corresponding DNA sequence of the locus. The dotted line highlights the coincident 5’ ends of the most abundant P4R2 RNAs at the locus (colored deep purple) and the most abundant siRNAs (colored black). (B) Heat map depicting the frequency of mismatched nucleotides at each position of RNAs ranging in size from 15 to 76 nt in dcl2 dcl3 dcl4 triple mutant plants. To correct for the frequency of errors inherent to sequencing, mismatch values for each position of 15–76 nt RNAs in wild-type plants were subtracted prior to plotting the data. Only read sequences with single mismatches or perfect matches to the reference genome were utilized for this analysis. (C) Over-expression and purification of recombinant RDR2. The image on the left shows a 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis(SDS-PAGE) gel, stained with Coomassie blue, showing molecular weight markers (M), proteins of un-infected High Five cells (lane 1), proteins of High Five cells 72 hr after infection with baculovirus expressing recombinant RNA-dependent RNA polymerase 2 (RDR2) (lane 2), and purified recombinant V5-tagged RDR2 after affinity purification and elution with V5 peptide. The image at right shows anti-RDR2 and anti-V5 immunoblots of the same three protein samples. For RDR2 detection, rabbit anti-RDR2 primary antibody was used in conjunction with donkey anti-rabbit HRP-conjugated secondary antibody. Detection of V5-tagged RDR2 involved anti-V5 HRP conjugate antibody. (D) RDR2 terminal transferase activity. Recombinant RDR2 or an active-site mutant form of RDR2 (RDR2-ASM) was incubated with alpha-labeled 32P-CTP and 51 nt RNA substrates bearing 3’ hydroxyl or 3’ dideoxy termini. Reaction products were subjected to denaturing polyacrylamide gel electrophoresis (PAGE) and autoradiography. For gel lane 4, reaction products were treated with RNase One, which degrades single-stranded RNAs, prior to PAGE. RNA size markers were run in lane M. The 51 nt RNA template, 5’ end-labeled using T4 polynucleotide kinase, was run as a size marker in the lane at far right.
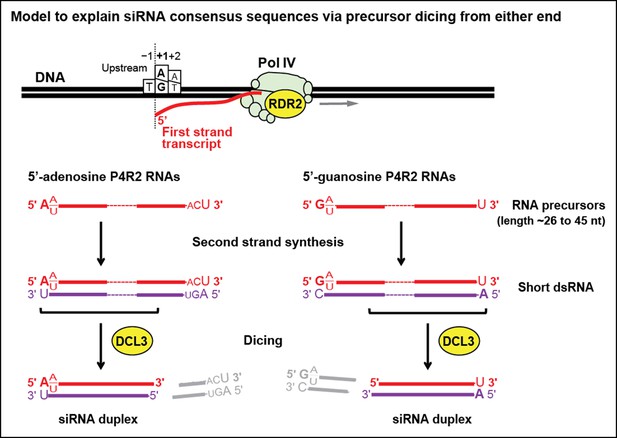
Model for the biogenesis of 24 nt small interfering RNAs (siRNAs) via single dicing events.
Pol IV/RDR2-dependent RNAs (P4R2 RNAs) tend to begin with a purine (A or G) at position 1, adjacent to what would be a T at position −1 in the corresponding DNA strand and an A (or U) at position 2. A similar signature is detected at the 5’ end of 24 nt siRNAs (see Figure 7), except that only A (not G) is enriched at the 5’ terminus of these siRNAs. Thus, DICER-like 3 (DCL3) cleavage measured from the 5’ adenosine of those P4R2 RNAs that begin with A could account for the similar 5’ end sequences of P4R2 RNAs and 24 nt siRNAs. P4R2 RNAs that begin with A tend to end with ACU, which is not reflected in siRNAs. However, P4R2 RNAs that begin with G tend to end with a 3’ U, with U > C > A being the order of preference, which matches the 3’ end consensus for 24 nt siRNAs. Because P4R2 RNAs that begin with G most often end with U, their complementary strands (purple bottom strands in the figure) would tend to have A at their 5’ ends. DCL3 dicing, measured from this 5’ A, would liberate a top strand (red in the figure) whose 3’ end could account for the 3’ consensus (U > C > A) of 24 nt siRNAs.
Tables
RNA sequencing statistics.
| Sample | Yield (Mb) | %PF | Cluster (PF) | Q30 | Mean qual. (PF) |
|---|---|---|---|---|---|
| Col-0 (Rep 1) | 2223 | 88.24 | 22,229,855 | 87.65 | 34.24 |
| dcl2/3/4 (Rep 1) | 2611 | 88.03 | 26,111,516 | 87.11 | 34.03 |
| nrpd1-3 (Rep 1) | 3220 | 89.06 | 32,204,710 | 89.20 | 34.75 |
| rdr2-1 (Rep 1) | 3500 | 88.60 | 34,995,990 | 87.89 | 34.33 |
| Col-0 (Rep 2) | 3345 | 88.36 | 33,446,115 | 87.87 | 34.32 |
| dcl2/3/4 (Rep 2) | 3029 | 88.27 | 30,291,115 | 88.49 | 34.50 |
| nrpd1-3 (Rep 2) | 3014 | 88.49 | 30,141,955 | 88.63 | 34.54 |
| rdr2-1 (Rep 2) | 3301 | 88.05 | 33,005,785 | 87.20 | 34.08 |
Mapping statistics.
| Sample | Total | Perfect match | % | Single mismatch | % |
|---|---|---|---|---|---|
| Col-0 (Rep 1) | 9,742,599 | 8,602,123 | 88.29 | 1,140,476 | 11.71 |
| dcl2/3/4 (Rep 1) | 8,440,663 | 7,179,444 | 85.06 | 1,261,219 | 14.94 |
| nrpd1-3 (Rep 1) | 8,966,872 | 7,799,160 | 86.98 | 1,167,712 | 13.02 |
| rdr2-1 (Rep 1) | 9,261,683 | 8,081,911 | 87.26 | 1,179,772 | 12.74 |
| Col-0 (Rep 2) | 13,955,193 | 12,179,431 | 87.28 | 1,775,762 | 12.72 |
| dcl2/3/4 (Rep 2) | 10,119,912 | 8,570,313 | 84.69 | 1,549,599 | 15.31 |
| nrpd1-3 (Rep 2) | 10,285,064 | 9,096,528 | 88.44 | 1,188,536 | 11.56 |
| rdr2-1 (Rep 2) | 9,970,701 | 8,793,492 | 88.19 | 1,177,209 | 11.81 |
Additional files
-
Supplementary file 1
Table showing the coordinates and normalized read counts for 24 nt small interfering RNA (siRNAs) and P4R2 RNAs detected in wild-type (Col-0), dcl2/3/4, pol IV and rdr2 within 100 bp windows.
Data for two independent RNA sequencing (RNA-seq) replicates for each genotype are provided.
- https://doi.org/10.7554/eLife.09591.018
-
Supplementary file 2
Pol IV/RDR2-dependent RNAs (P4R2 RNAs) overlap with Pol IV-dependent transcript loci identified by Li et al, 2015.
P4R2 RNAs detected in the dcl2 dcl3 dcl4 (dcl2/3/4) triple mutant were compared to a set of 22 Pol IV-dependent transcript loci verified by reverse transcription-polymerase chain reaction (RT-PCR) in the study of Li et al., 2015. The number of P4R2 RNAs ≥ 26 nt overlapping each Pol IV locus is shown for dcl2/3/4 replicate 1 and 2 RNA sequencing (RNA-seq) datasets. Unique reads are those that map to only one genomic location. Total reads include reads that can map to two or more genomic loci.
- https://doi.org/10.7554/eLife.09591.019





