Sir2 phosphorylation through cAMP-PKA and CK2 signaling inhibits the lifespan extension activity of Sir2 in yeast
Figures
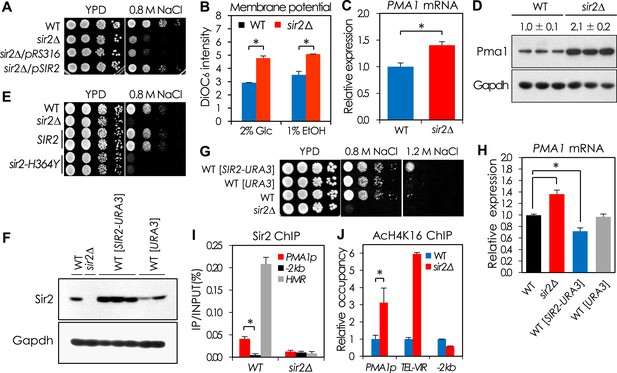
Sir2 negatively regulates PMA1 expression by deacetylating H4K16 in the PMA1 promoter.
(A) NaCl sensitivity of wild-type (WT), sir2∆, and sir2∆ cells expressing SIR2. (B) Plasma membrane potential as indicated by DiOC6 staining of WT and sir2∆ cells grown in glucose or ethanol medium (*p < 0.005). (C) PMA1 mRNA levels in WT and sir2∆ cells measured by qRT-PCR (*p < 0.001). (D) Pma1 protein levels in WT and sir2∆ cells measured by Western blot (WB). (E) NaCl sensitivity of WT, sir2∆, and sir2∆ cells carrying WT SIR2 or the sir2-H364Y mutant allele. (F) Silent information regulator 2 (Sir2) protein levels of the SIR2-overexpressing cells measured by WB. (G) NaCl sensitivity of the SIR2-overexpressing cells. (H) PMA1 mRNA levels of the SIR2-overexpressing cells measured by qRT-PCR (*p < 0.02). (I) Sir2 enrichment at the PMA1 promoter measured by Sir2 ChIP (*p < 0.001). (J) H4K16 acetylation levels at the PMA1 promoter in WT and sir2∆ cells measured by AcH4K16 ChIP (*p < 0.02). Values in (B), (C), (H), (I), and (J) represent the average of at least three independent experiments (±S.D.).
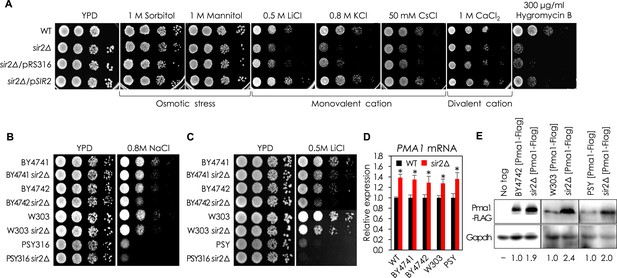
Sir2 is involved in regulation of monovalent cations in yeast.
(A) sir2∆ mutant was sensitive to various monovalent cations and Hygromycin B, but not to divalent cation or osmotic stress, than WT. WT, sir2∆, sir2∆ cells transformed with an empty low-copy vector (pRS316) or pRS316 carrying the SIR2 gene were spotted onto each plate as indicated and incubated at 30°C for 2 days. (B, C) sir2∆ mutants of all strains tested were more sensitive to NaCl (B) and LiCl (C) than WT in solid medium. (D) PMA1 mRNA levels in sir2∆ mutants were higher than those in WT strains (*, vs WT in an indicated strain, p < 0.03). The values are the average of at least three independent experiments (±S.D.). (E) Pma1 protein levels in WT strains and their sir2∆ mutants were analyzed by Western blotting using an anti-Flag antibody. GAPDH protein was used as a loading control.
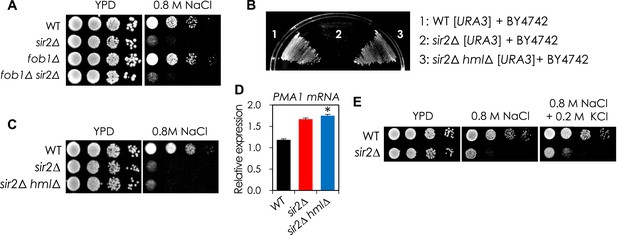
The NaCl sensitivity of the sir2Δ mutant was affected neither by Fob1 nor by pseudodiploid state.
(A) Deletion of FOB1 gene does not affect NaCl sensitivity in WT and sir2∆. Each cell was spotted onto YPD plates with 0.8 M NaCl and incubated at 30°C for 3 days. (B) Deletion of HMLα from the sir2∆ mutant restored the mating phenotype. Each of mating type a cells (WT [URA3], sir2∆ [URA3], and sir2∆ hml∆ [URA3]) and α cells (BY4742) was seeded together into YPD medium at an initial OD600 = 0.2 and then incubated at 30°C for 1 day. Five μl of the cell culture was collected and spread onto synthetic minimal plate containing leucine and histidine. (C) Deletion of HMLα does not affect NaCl sensitivity in sir2∆ mutant. (D) Deletion of HMLα does not affect PMA1 mRNA level in sir2∆ mutant. The PMA1 mRNA level of each cell was analyzed by qRT-PCR (*, vs WT in an indicated strain, p < 0.005). (E) Addition of KCl did not rescue the NaCl sensitivity of sir2∆ cells. Each cell was spotted onto YPD plates containing 0.8 M NaCl with or without 0.2 M KCl and incubated at 30°C for 2 days.
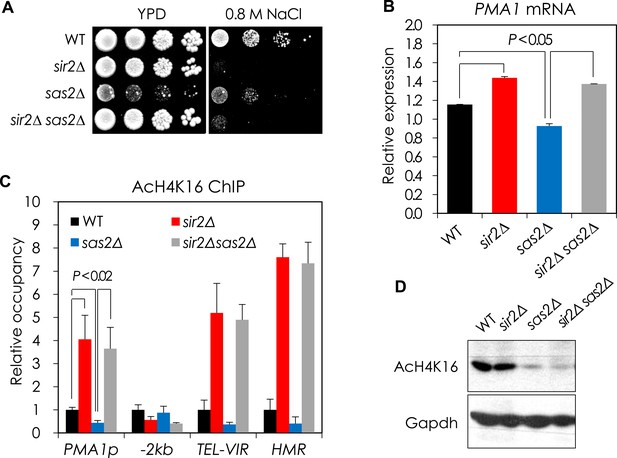
Sir2 and Sas2 antagonistically regulate PMA1 transcription in part through regulating H4K16 acetylation at PMA1 promoter.
(A) NaCl sensitivity of WT, sir2∆, sas2∆, and sir2∆ sas2∆ cells. Each cell was spotted onto YPD plates containing 0.8 M NaCl and incubated at 30°C for 3 days. (B) The PMA1 mRNA levels of WT, sir2∆, sas2∆, and sir2∆ sas2∆ cells were analyzed by qRT-PCR. ACT1 was used as a loading control. The values are the average of at least three independent experiments (±S.D.). p values were calculated using t-test. (C) Changes in H4K16 acetylation at the PMA1 promoter were assayed by chromatin immunoprecipitation (ChIP) using an anti-H4K16 antibody in WT, sir2∆, sas2∆, and sir2∆ sas2∆ cells. 2 kb upstream region of the PMA1 promoter (−2 kb) was used as a negative control for Sir2 action. TEL-VIR and HMR are known Sir2 targets. The values are the average of at least three independent experiments (±S.D.). p values were calculated using t-test. (D) Total H4K16 acetylation levels were analyzed by Western blotting using the anti-H4K16 antibody in WT, sir2∆, sas2∆, and sir2∆ sas2∆ cells. GAPDH protein was used as a loading control.
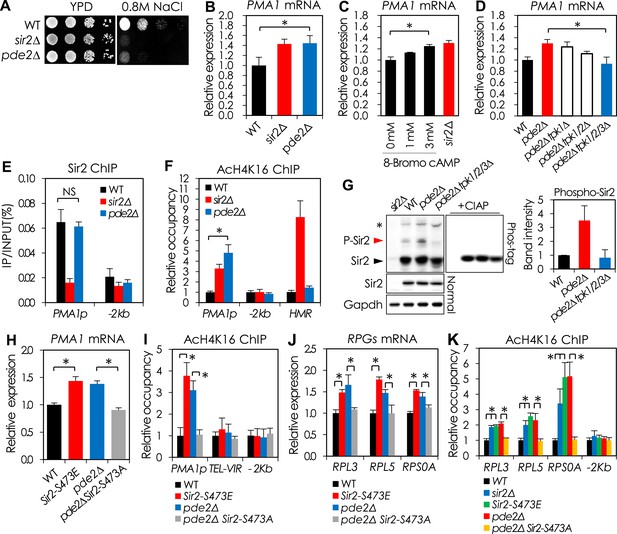
cAMP-PKA signaling inhibits Sir2 activity for the transcriptional repression of PMA1 and RPGs through serine 473 phosphorylation.
(A) Effects of PDE2 deletion on NaCl sensitivity. (B) Effects of PDE2 deletion on PMA1 expression measured by qRT-PCR (*p < 0.01). (C) Effects of 8-Bromo-cAMP addition on PMA1 expression measured by qRT-PCR (*p < 0.01). (D) Effects of TPK deletion on PMA1 expression in pde2∆ cells measured by qRT-PCR (*p < 0.01). (E) Sir2 enrichment at the PMA1 promoter in WT and pde2∆ cells measured by Sir2 ChIP (NS, not significant). (F) H4K16 acetylation levels at the PMA1 promoter in WT, sir2∆, and pde2∆ cells measured by AcH4K16 ChIP (*p < 0.001). (G) Sir2 phosphorylation levels in WT, pde2∆, and pde2∆ tpk1/2/3∆ cells analyzed by Phos-tag SDS-PAGE and WB. Arrowheads indicate cAMP-PKA-dependent phosphorylated (red) and non-phosphorylated (black) Sir2. The asterisk indicates cAMP-PKA-independent phosphorylation of Sir2. (H) Effects of SIR2-S473E or SIR2-S473A on PMA1 expression in WT and pde2∆ cells measured by qRT-PCR (*p < 0.005). (I) Effects of SIR2-S473E or SIR2-S473A on H4K16 acetylation at the PMA1 promoter in WT and pde2∆ cells measured by AcH4K16 ChIP (*p < 0.05). (J) Effects of SIR2-S473E or SIR2-S473A on the expression of ribosomal protein genes (RPGs) in WT and pde2∆ cells measured by qRT-PCR (*p < 0.005). (K) Effects of SIR2-S473E or SIR2-S473A on H4K16 acetylation at the RPG promoters in WT and pde2∆ cells measured by AcH4K16 ChIP (*p < 0.05). Values in (B), (C), (D), (E), (F), (H), (I), (J), and (K) represent the average of at least three independent experiments (±S.D.).
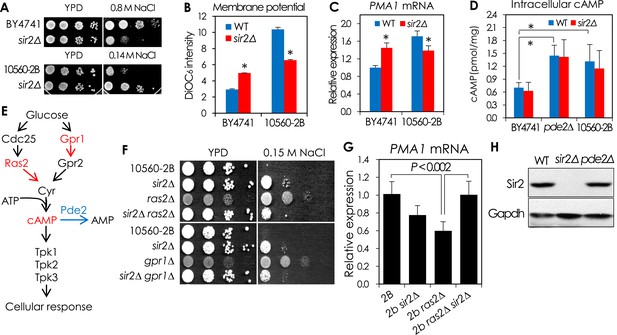
The cAMP-PKA signaling-dependent effects of SIR2 deletion on NaCl sensitivity and PMA1 expression.
(A–D) The effect of SIR2 deletion on NaCl sensitivity and PMA1 expression in a sigma background 10560-2B strain is opposite to that in BY4741. The 10560-2B strain has hyperactive cAMP-PKA pathway, which confers an increased NaCl sensitivity to the strain. (A) NaCl sensitivity of WT and sir2∆ in BY4741 and 10560-2B. (B) Plasma membrane potential of WT and sir2∆ in BY4741 and 10560-2B (*, vs WT in an indicated strain, p < 0.005). (C) PMA1 mRNA levels of WT and sir2∆ in BY4741 and 10560-2B measured by qRT-PCR (*, vs WT in an indicated strain, p < 0.0001). (D) Intracellular cAMP levels of WT and sir2∆ in BY4741, BY4741 pde2∆ and 10560-2B (*p < 0.005). (E–H) Reduction of cAMP/PKA signaling reversed the effect of SIR2 deletion on the PMA1 expression in 10560-2B strain. (E) Overview of the cAMP/PKA signaling pathway. (F) Effect of deleting either RAS2 or GPR1, encoding an activator of adenylate cyclase or a G-protein glucose receptor, on the NaCl sensitivity of WT 10560-2B and sir2∆ cells. (G) The PMA1 mRNA levels of WT 10560-2B, sir2∆, ras2∆, and sir2∆ ras2∆ cells were analyzed by qRT-PCR. (H) Sir2 protein levels in WT and pde2∆ cells measured by WB. GAPDH protein was used as a loading control.
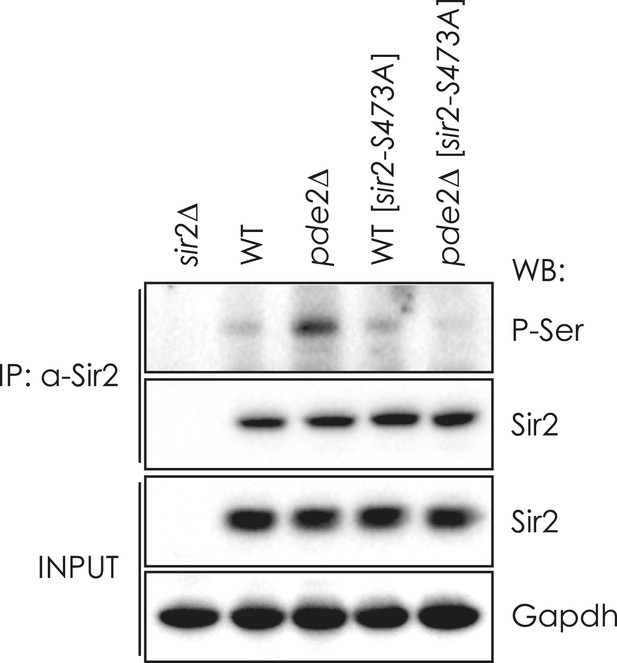
Effect of sir2-S473A mutation on the phosphorylation of Sir2 in WT and pde2∆ cells.
Sir2 proteins in WT, sir2∆, pde2∆, WT [sir2-S473-A], and pde2∆ [sir2-S473A] cells were IP with anti-Sir2 antibody and analyzed by WB as indicated. GAPDH protein was used as a loading control.
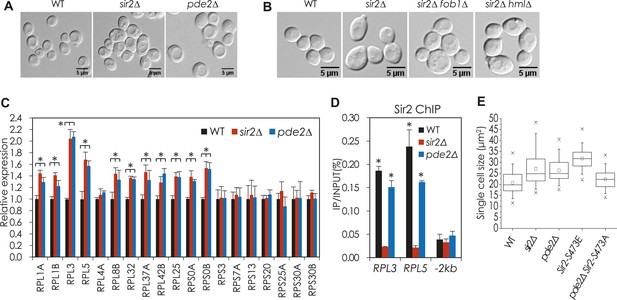
cAMP-PKA signaling and phosphorylation of Sir2 at S473 regulate the expression of genes encoding ribosomal subunit proteins and cell size homeostasis.
(A) Cell size of WT, sir2∆, and pde2∆ cells. Cells grown in YPD were harvested at an OD600 = 0.5. (B) Cell size of WT, sir2∆, sir2∆ fob1∆, and sir2∆ hml∆ cells. Cells grown in YPD were harvested at an OD600 = 0.5. (C) mRNA levels of RPGs in WT, sir2∆, and pde2∆ cells (*, vs WT, p < 0.005). The values are the average of at least three independent experiments (±S.D.). (D) Sir2 enrichment at the RPL3 and RPL5 promoters in WT, sir2∆, and pde2∆ cells (*, vs WT, p < 0.01). 2 kb upstream region of the PMA1 promoter (−2 kb) was used as a negative control for Sir2 action. The values are the average of at least three independent experiments (±S.D.). (E) Effect of SIR2-S473E and SIR2-S473A on single cell size of WT and pde2∆ cells. Values correspond to the average single cell sizes measured for four independent cultures (>150 cells for each culture).
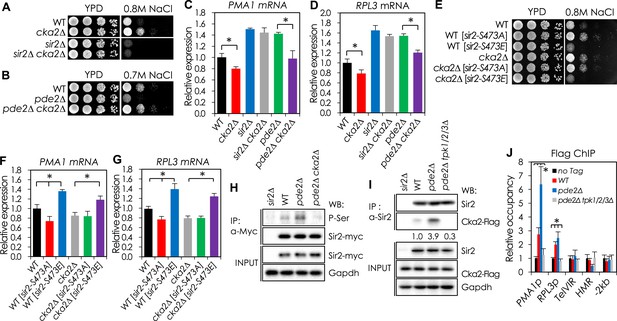
CKA2 mediates cAMP-PKA-dependent Sir2 phosphorylation to regulate the expression of PMA1 and RPGs.
(A, B) Effects of CKA2 deletion on NaCl sensitivity in WT (A), sir2∆ (A), and pde2∆ cells (B). (C, D) Effects of CKA2 deletion on the expression of PMA1 (C) and RPL3 (D) in WT, sir2∆, and pde2∆ cells measured by qRT-PCR (*p < 0.01). (E) Effects of SIR2-S473E or SIR2-S473A on NaCl sensitivity in WT and cka2∆ cells. (F, G) Effects of SIR2-S473E or SIR2-S473A on the expression of PMA1 (F) and RPL3 (G) in WT and cka2∆ cells measured by qRT-PCR (*p < 0.005). (H) Sir2 phosphorylation levels in WT, pde2∆, and pde2∆ cka2∆ cells. Myc-tagged Sir2 proteins were immunoprecipitated (IP) and analyzed by WB as indicated. (I) In vivo Sir2 and Cka2 interaction in WT, pde2∆, and pde2∆ tpk1/2/3∆ cells. Flag-tagged Cka2 proteins (Cka2-Flag) were IP and analyzed by WB. (J) Cka2-Flag enrichment at the PMA1 promoter in WT, pde2∆, and pde2∆ tpk1/2/3∆ cells measured by Flag ChIP (*p < 0.001). Values in (C), (D), (F), (G), and (J) represent the average of at least three independent experiments (±S.D.). NaCl sensitivity of the 121 kinase mutant strains used to identify kinases required for protein kinase A (PKA)-dependent Sir2 phosphorylation is available in the Figure 3—source data 1.
-
Figure 3—source data 1
NaCl sensitivity of the kinase mutant strains.
- https://doi.org/10.7554/eLife.09709.012
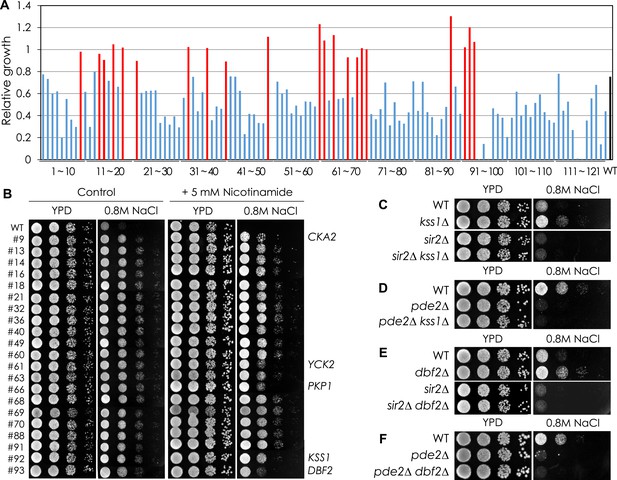
Sir2-mediated NaCl sensitivity of kinase mutant strains.
(A) NaCl sensitivity of kinase mutant strains. (B) Effect of nicotinamide treatment on the NaCl sensitivity of 21 kinase mutants. Each cell was spotted onto YPD plates containing 0.8 M NaCl with or without 5 mM nicotinamide and incubated at 30°C for 2 days. (C, D) Effect of KSS1 deletion on the NaCl tolerance in WT (C), sir2∆ (C), and pde2∆ (D). (E, F) Effect of DBF2 deletion on the NaCl tolerance in WT (E), sir2∆ (E), and pde2∆ (F).
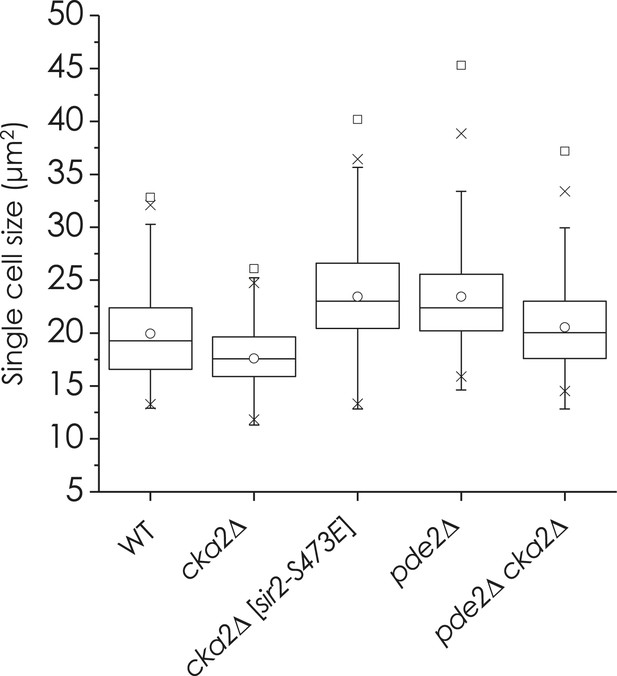
Effect of cka2∆ or sir2-S473E mutation on cell size of WT and pde2∆ mutant cells.
WT, cka2∆, cka2∆ [sir2-S473E], pde2∆, and pde2∆ cka2∆ cells were grown in YPD and harvested at an OD600 = 0.5. Cell sizes were measured for at least 250 unbudded single cells.
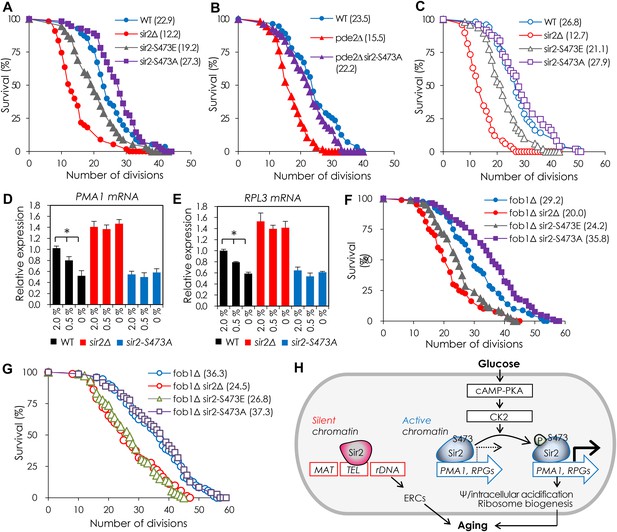
Sir2 S473 phosphorylation inhibits DR-mediated lifespan extension by Sir2.
(A) Replicative lifespan (RLS) of the strains expressing SIR2-S473E or SIR2-S473A measured by micromanipulation. The median lifespan is indicated. p < 0.0001 (WT vs sir2Δ), p = 0.0013 (WT vs sir2-S473E), p = 0.0062 (WT vs sir2-S473A). (B) Effect of SIR2-S473A on the RLS of pde2∆ cells. The median lifespan is indicated. p < 0.0001 (WT vs pde2Δ), p < 0.0001 (pde2Δ vs pde2Δ sir2-S473A). (C) RLS of WT, sir2∆, and strains expressing SIR2-S473E or SIR2-S473A grown under 0.5% glucose conditions. The median lifespan is indicated. p < 0.0001 (WT vs sir2Δ), p = 0.0006 (WT vs sir2-S473E), p = 0.5051 (WT vs sir2-S473A). (D, E) Effects of SIR2-S473A on the expression of PMA1 (D) and RPL3 (E) in cells grown under 2.0%, 0.5%, or 0% glucose conditions measured by qRT-PCR (*p < 0.005). The values represent the average of at least three independent experiments (±S.D.). (F, G) Effects of SIR2-S473E or SIR2-S473A on the RLS of fob1∆ background under 2% (F) and 0.5% (G) glucose conditions. The median lifespan is indicated. p < 0.0001 (fob1∆ vs sir2Δ under 2.0% glucose), p < 0.0001 (fob1∆ vs sir2-S473E under 2.0% glucose), p = 0.0002 (fob1∆ vs sir2-S473A under 2.0% glucose), p < 0.0001 (fob1∆ vs sir2Δ under 0.5% glucose), p < 0.0001 (fob1∆ vs sir2-S473E under 0.5% glucose), p = 0.3221 (fob1∆ vs sir2-S473A under 0.5% glucose). (H) A working model for how Sir2 regulates dietary restriction (DR)-mediated lifespan in yeast.
Additional files
-
Supplementary file 1
Strains used in the study.
- https://doi.org/10.7554/eLife.09709.016
-
Supplementary file 2
Primers used in the study.
- https://doi.org/10.7554/eLife.09709.017





