Dna2 nuclease-helicase structure, mechanism and regulation by Rpa
Figures
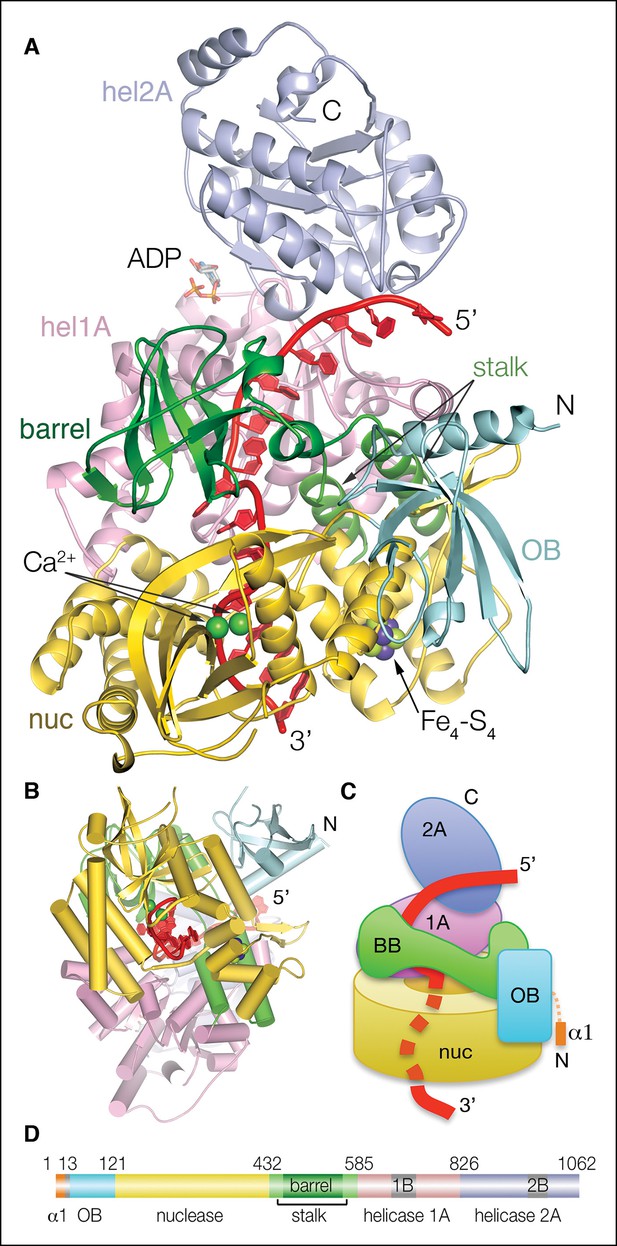
Structure of the Dna2-ssDNA complex.
(A) Cartoon representation of the Dna2-ssDNA complex. The α1 helix, which packs with the hel2A domain of a crystallographic symmetry related protomer, is omitted from this view. The individual domains of Dna2 are colored separately as indicated in (D), ssDNA is red, ADP is shown as sticks, calcium ions are shown as green spheres, the iron-sulfur cluster is shown in a space-filling representation (nuc: nuclease domain; hel1A: helicase 1A domain, hel2A: helicase 2A domain, OB: oligonucleotide/oligosaccharide binding domain). (B) View looking up the vertical axis of (A). (C) Schematic of the complex illustrating the relative arrangement of the Dna2 domains, and highlighting its cylinder-like shape. Colored as in (A). (D) Linear representation of the Dna2 domains and their boundaries; colored as in (A).
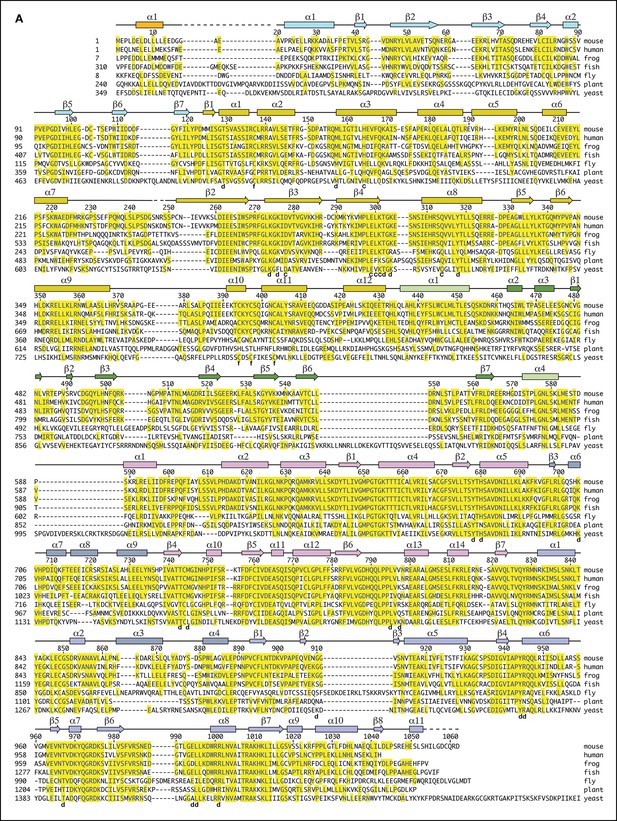
Dna2 secondary structure and sequence conservation.
Helices are indicated as cylinders, b strands as arrows, segments lacking regular secondary structure as solid lines, and disordered regions as dashed lines. Secondary structure elements are colored as in Figure 1A. The N-terminal extensions, relative to mammalian Dna2, of the other orthologs are not shown. In the alignment of yeast Dna2, reliable homology starts with the β2 strand of the OB domain at residues 44 and 417 of the mouse and yeast orthologs, respectively. The ~360 residue, unique N-terminal segment of yeast Dna2 has an overall low hydrophobicity content indicative of lack of globular structure, and short regions of homology with closely related yeasts suggests that this region acts through linear epitopes, some of which may be acidic/amphipathic helices (not shown). Residues that contact the DNA are marked by “d”, active site residues by “c” and iron-sulfur cluster ligands by “f”. The entire protein structure is within 0.9 Å Cα r.m.s.d. in DNA-bound and apo Dna2 crystals, and the overall DNA conformation is essentially identical in a different crystal form of Dna2 bound to a 21-nt ssDNA (Table 1).
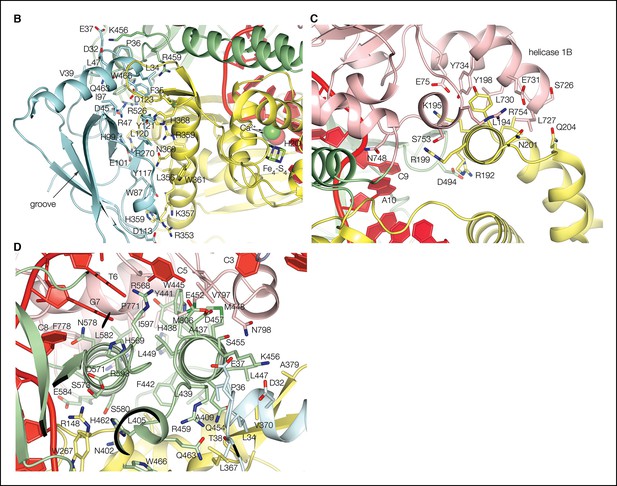
Dna2 inter-domain interfaces.
(A) The nuclease domain packs with the OB domain extensively, through both polar and hydrophobic residues. The four residues between the last b strand of the OB domain and the first b strand of the nuclease domain are buried in the interface between the two domains. The OB groove, where DNA or peptides bind in other OB fold proteins, is solvent exposed opposite from its surface that packs with the nuclease domain. Individual domains are colored as in Figure 1A. (B) The Dna2 nuclease core fold has a three-helix insertion (residues 176 to 227) that packs with the helicase 1B domain. This three-helix insertion is also present in the AddB nuclease, where it packs with the N-terminal portion of the protein. (C) The nuclease domain also links to the helicase domain indirectly, through the stalk domain that is sandwiched between the two.
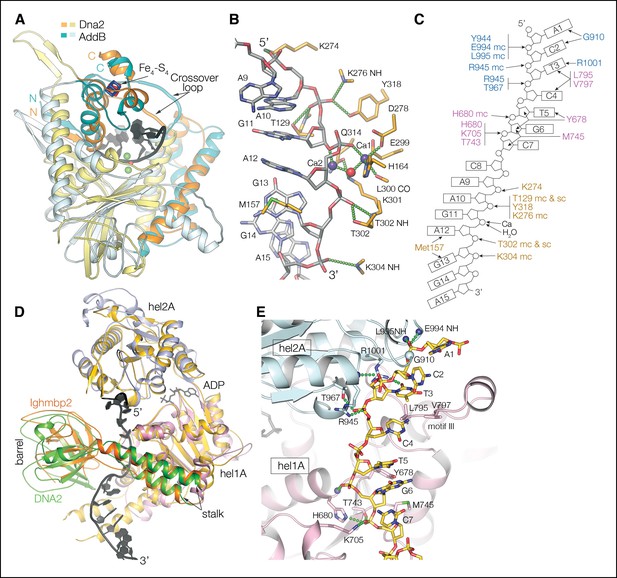
Nuclease and helicase domain structures and DNA contacts.
(A) Superposition of the Dna2 nuclease domain on the AddB nuclease domain. The N-terminal βαα extension (residues 122 to 154) and the C-terminal αα extension (residues 384 to 412) is colored in orange, with the corresponding elements of AddB in dark cyan. Green spheres are calcium ions. (B) DNA contacts and active site residues of the Dna2 nuclease domain. Hydrogen bonds are depicted as green dotted lines, calcium ions as blue spheres, water as a red sphere. (C) Diagram showing the contacts depicted in (B) and (E). The residues are colored according to the domain they belong as in Figure 1D (mc: main chain, sc: side chain). (D) Superposition of the Dna2 helicase domain on Ighmbp2. Dna2 is colored as in Figure 1D. The Ighmbnp2 1A (hel1A) and 2A (hel2A) helicase domains are colored gold, its β barrel in light orange, and stalk dark orange. (E) DNA contacts of the helicase 1A (pink) and 2A (cyan) domains, showing residues that are involved in either hydrogen bond (green dotted lines) or van der Waals contacts.
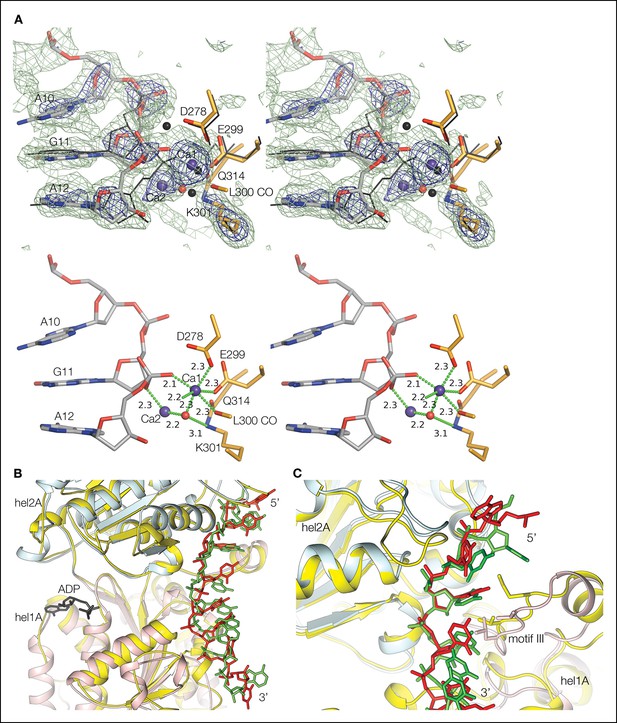
Electron density at the nuclease active site and structural similarity of the Dna2 helicase to Upf1-subfamily RNA/DNA helicases.
(A) Top panel shows a stereo view of the mFo-DFc electron density at the active site, calculated with phases after omitting the two calcium ions (blue spheres), the water molecule (red sphere), a 3-nt segment centered on the scissile phosphate group and the side chain of Lys301, and subjecting the rest of the entire structure to simulated annealing from 3000°K. The dark blue map is contoured at 5 σ, and the light green map at 2.5 σ. The orientation is similar to that of Figure 2B. For comparison, the corresponding residues of the λ nuclease-DNA complex (Zhang et al., 2011) are shown as black sticks (the lysine is mutated to alanine in that structure), and the calcium ions and water molecule as black spheres. The Dna2 Ca-1 ion and water molecule have a coordination shell and position very similar to the first magnesium and associated water in λ nuclease, but the position of Ca-2 is different. The Ca coordination shell and residue labels are shown in bottom panel. For clarity, not shown are the electron density, λ nuclease, and the His164 side chain to which the green-dotted bond from Ca-1 would be connected. (B) Superposition of Dna2 helicase domains 1A (pink) and 2A (cyan) with Ighmbp2 (gold). DNA is colored in red, RNA is colored in green. The only regions of the Dna2 helicase that do not have counterparts in the Upf1 subfamily are the ~40 residue 1B domain, which packs with the nuclease domain, and the ~20 residue 2B domain, which is distal from the rest of the protein and the DNA (Figure 1D). (C) Superposition of individual Dna2 helicase domains 1A (pink) and 2A (cyan) with Upf1 (gold). DNA is colored in red, RNA is colored in green. Motif III residues are shown as sticks. Supplemental discussion of helicase translocation. Based on the proposed SF1B translocation mechanism (Saikrishnan et al., 2009), ATP binding and concomitant closure of the cleft between the 1A and 2A domains would be coupled to domain 2A releasing the DNA and rebinding it at a register shifted by 1 nt in the 5’ to 3’ direction, while domain 1A would retain its DNA. On ATP hydrolysis, domain 1A would be the one releasing its DNA and rebinding it at the +1 nt register, restoring the initial open-cleft conformation. In the Dna2-ssDNA structure, however, the rebinding of 1A to DNA at the shifted register will have to await the transient dissociation of the nuclease domain from DNA. The increased half life of the closed cleft would likely also reduce the rate of exchange of ADP for ATP, as the γ phosphate binding site of the helicase fold is far less solvent accessible in the closed cleft.
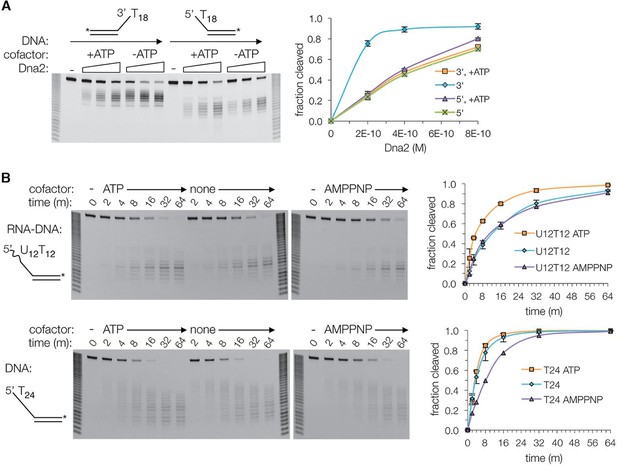
Dna2 nuclease activity.
(A) Denaturing PAGE showing ATP inhibits the nuclease activity on 3’ overhang substrate, while slightly increasing it for the 5’ overhang substrate. Substrates are at 15 nM. For this and subsequent nuclease assays, cleavage was quantified by loss of substrate and plotted with ± s.d. error bars (n = 3). (B) Nuclease time course of 2 nM Dna2 with 10 nM 5’ RNA-DNA overhang or 5’ DNA overhang substrates. ATP or AMPPNP is at 1.3 mM.
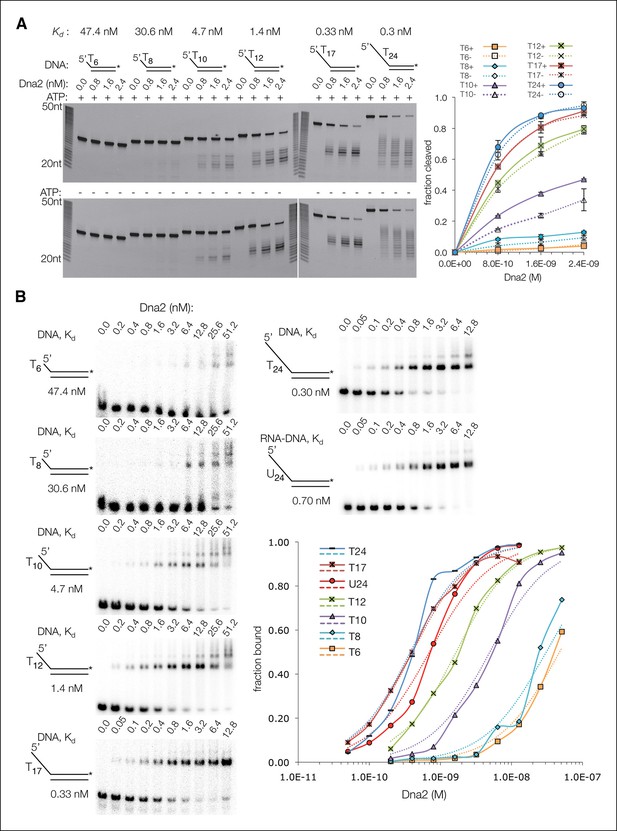
The ssDNA-length dependence of the DNA affinity correlates with cleavage rates.
(A) Denaturing PAGE showing length dependency and cleavage sites of 5’ overhang DNA at a 30 nM concentration (asterisk indicates FAM labeling of the 3’ end) in the presence (top panel) or absence (bottom) of 1.3 mM ATP. Also shown are the dissociation constants (Kd) of Dna2(D278A) for each substrate, according to the data shown in Figure 3—figure supplement 1B. (B) Electrophoretic mobility shift assays (EMSA) of Dna2(D278A) binding to 5’ overhang DNA substrates. The graph shows fraction bound. Dotted lines are simulated binding curves based on the indicated Kd values.
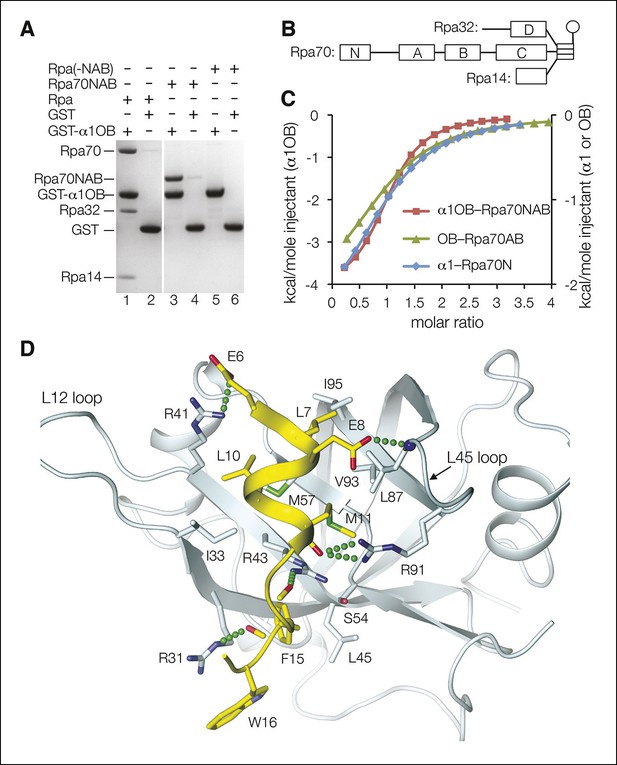
Dna2 physically interacts with Rpa.
(A) GST pull-down assay showing that α1OB of Dna2 (residues 1–122) interacts with Rpa70NAB (1–431), but not the rest of Rpa heterotrimer (Rpa(-NAB)). (B) Schematic drawing of the Rpa trimer, showing the OB domains as rectangles and the winged helix (WH) domain of Rpa32 as a sphere. (C) ITC curves for the human α1OB-Rpa70NAB, α1-Rpa70N and OB-Rpa70AB complexes. (D) Structure of the α1-Rpa70N complex. The α1 peptide is in yellow and Rpa70N in cyan. For clarity, the main chain amide group of L87 and carbonyl groups of M11, K13, F15 are not labeled.
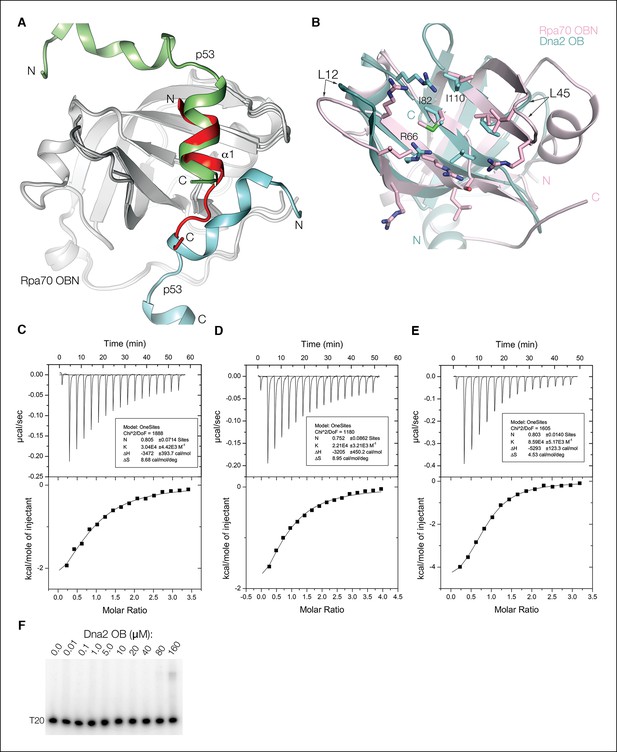
Comparison of the Dna2 α1–Rpa70 OBN, p53–Rpa70 OBN and Dna2 OB structures.
(A) Superposition of the α1–Rpa70 OBN structure on the fusion protein of Rpa70 OBN and p53. In the fusion protein structure, the Rpa70 OBN (gray) groove has two p53 peptides, one (cyan) from its fused p53 and another (green) from a crystallographic-symmetry related fusion protein. While it was suggested that only the latter represented the solution-state complex, our structure has aspects of both p53-Rpa70 OBN interfaces. An acidic/amphipathic helix from the symmetry-related p53 superimposes with the Dna2 helix, and part of the second p53 peptide overlaps with the Dna2 β turn and associated contacts. α1 peptide is red. (B) The Dna2 OB groove (cyan) has an overall hydrophobic- and basic-residue content similar to that of the Rpa70 OBN (pink). The L12 and L45 loops (labeled), which in DNA-binding OB domains partially wrap around the DNA, but are truncated in the Rpa70 OBN domain are, respectively, truncated and absent in the Dna2 OB domain, resulting in a wide open groove. Residues mutated in Figure 5A are labeled. (C) ITC titration profile of human Dna2 α1 (residues 1–20) binding to human Rpa70N (residues 1–120). (D) ITC titration profile of human Dna2 OB (residues 21–122) binding to human Rpa70AB (residues 181–422). (E) ITC titration profile of human Dna2 α1OB (residues 1–122) binding to human Rpa70NAB (residues 1–422). (F) The Dna2 OB domain does not exhibit any detectable DNA-binding at concentrations up to 80 μM in an EMSA assay with (dT)20 ssDNA. A shifted, smeary band appearing at 160 μM OB has ~ 4% of the total DNA.
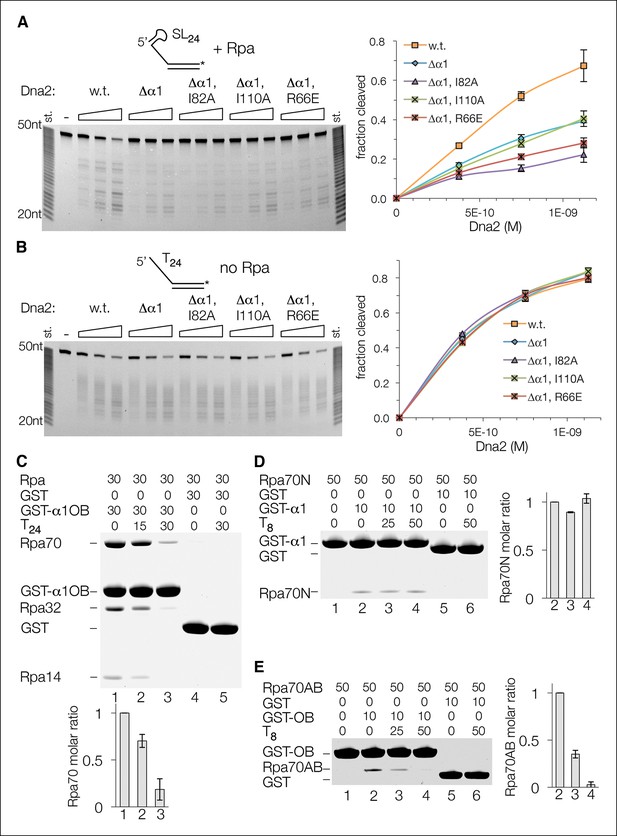
Both Dna2-Rpa interactions are important for Dna2 stimulation, but only one is mutually exclusive with Rpa-ssDNA interactions.
(A) Cleavage of a 5’ stem-loop overhang substrate (15 nM) by wild type, α1-deleted, and OB mutant Dna2 in the presence of 15 nM Rpa. (B) Nuclease activity of the same set of enzymes as in (A), but using a 5’ (dT)24 overhang substrate in the absence of Rpa. (C) GST pull-down assay showing that α1OB and (dT)24 ssDNA bind to Rpa in a partially mutually exclusive manner. Protein and DNA concentrations are in μM, and the bar graph shows the quantitation of Rpa70 binding relative to the lane in the absence of DNA, which is set to 1. Error bars are standard deviations from three repetitions of each experiment. (D) The binding of α1 to the Rpa70N polypeptide is unaffected by (dT)8 ssDNA. GST pull-down assay and quantitation as in (C). (E) The binding of OB to the Rpa70AB polypeptide is abolished by (dT)8 ssDNA. GST pull-down assay and quantitation as in (C).
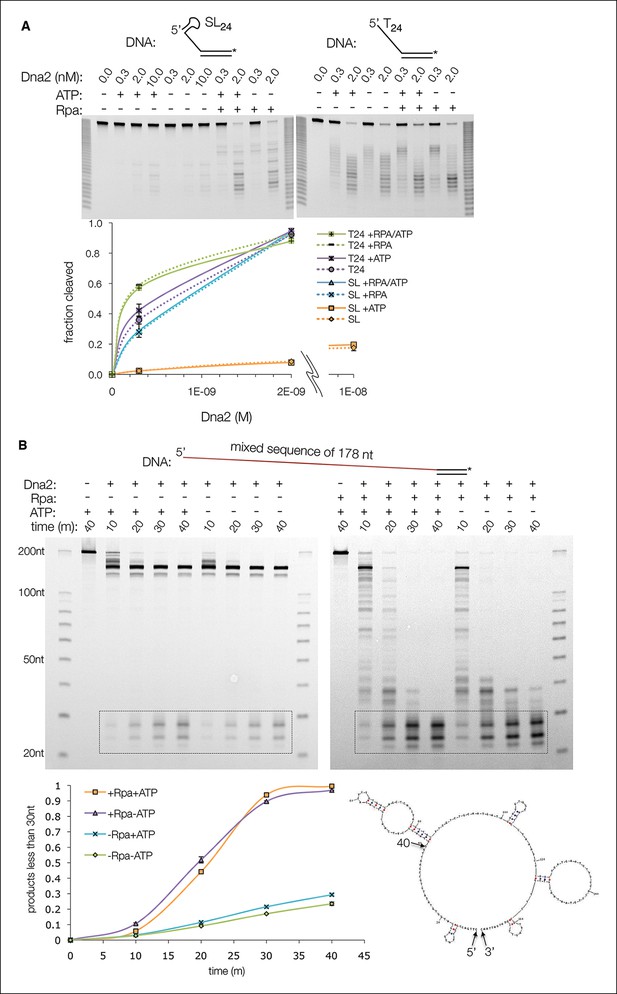
Cleavage of a ssDNA containing secondary structure is Rpa dependent.
(A) Cleavage of a 5’ overhang substrate that has a predicted 6 bp stem-loop secondary structure at the 5’ end of the ssDNA (5’ SL24). Dna2, at the indicated concentrations, failed to cleave 5’ SL24 (30 nM) appreciably under conditions where ~95% of the corresponding 5’ (dT)24 substrate was cleaved, irrespective of ATP. Addition of Rpa, at a stoichiometric amount to ssDNA, stimulated SL24 cleavage by over a factor of 20, but had only a modest effect on (dT)24 cleavage. In the presence of Rpa, the cleavage of the two substrates was within a factor of two, compared to being nearly two orders of magnitude apart in its absence. (B) Cleavage of a 178-nt long 5’ overhang substrate (80 nM) by Dna2 (20 nM). As with the shorter 5’ overhang substrates, ATP (1.3 mM) has only a minor stimulatory effect. Because mixed-sequence ssDNA of this length would invariably have some secondary structure (the lowest energy secondary structure, predicted by the UNAfold server (mfold.rna.albany.edu), is shown at bottom, right), cleavage is stimulated by Rpa (400 nM) as with the 5’ SL24 substrate. For this specific experiment, samples are quantified by integration of final products in the box.
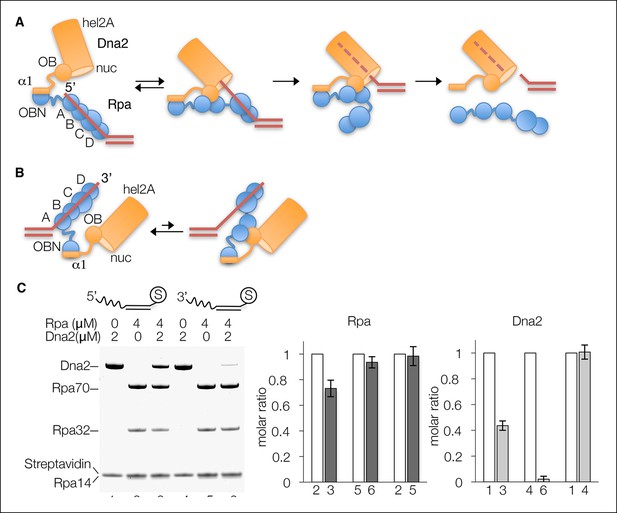
Dna2 displaces Rpa from 5’ but not 3’ overhang DNA.
(A) Schematic of the proposed mechanism of Dna2 displacing Rpa from a 5’ overhang DNA. Dna2 is shown as a hollow cylinder, except for its α1 helix and OB fold domains that are shown as a rectangle and circle, respectively. The label “nuc” marks the nuclease tunnel entrance into which the ssDNA would thread, and “hel2A” marks the helicase tunnel exit where the 5’ end of the ssDNA would end up after threading. (B) Schematic illustrating that at a 3’ overhang DNA, Dna2-Rpa interactions do not result in a free ssDNA end that can thread into the Dna2 tunnel. (C) Nuclease-dead Dna2 (D278A) displaces Rpa from 5’- but not 3’ overhang DNA. The overhang consists of (dT)26 and the DNA is conjugated to streptavidin (shown by “S”) through a biotin group at the end of the duplex. Column graph showing quantitation of Rpa32 plots the molar ratio relative to the reaction lacking Dna2 for each DNA substrate (lanes 2, 3 for 5’ overhang DNA, and lanes 5 and 6 for 3’ overhang DNA), or relative to lane 1 for the comparison of Rpa loading onto 5’- and 3’-overhang DNA. Quantitation of relative Dna2 loading is similarly shown in the last column graph. Error bars are standard deviations from three repetitions of each experiment.
Tables
Data collection and refinement statistics.
| Data Set | Dna2-5’ overhang DNA* | Dna2-ssDNA | apo Dna2 | apo Dna2 (SeMet) | DNA2 α1-RPA 70N |
|---|---|---|---|---|---|
| Space group | P22121 | P212121 | P212121 | P212121 | C121 |
| a, b, c (Å) | 87.2, 118.5, 149.3 | 120.2, 149.2, 172.9 | 120.9, 148.6, 170.5 | 120.9, 148.6, 170.5 | 134.3, 50.9, 76.5 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 103.9, 90.0 |
| Resolution (Å) | 50.0 - 2.35 (2.43 - 2.35) | 30.0 – 3.11 (3.23 - 3.11) | 50.0 – 3.0 (3.11 - 3.0) | 30.0 – 3.4 (3.52 - 3.4) | 60.0 – 1.5 (1.55 - 1.5) |
| Rsym | 12.6 (65.6) | 13.7 (87.5) | 13.1 (78.7) | 15.8 (59.3) | 7.4 (80.1) |
| Rpim | 6.3 (36.6) | 6.6 (57.1) | 6.8 (41.9) | 4.0 (15.1) | 5.1 (54.1) |
| I/σ(I) | 16.4 (1.9) | 13.2 (1.4) | 7.9 (1.3) | 19.3 (4.7) | 23.5 (3.2) |
| Completeness (%) | 99.0 (98.6) | 99.0 (99.2) | 98.3 (98.9) | 100.0 (100) | 84.3 (79.7) |
| Redundancy | 5.0 (3.9) | 6.1 (6.0) | 4.4 (4.5) | 15.1 (15.3) | 2.7 (2.7) |
| Refinement | |||||
| Resolution (Å) | 50.0-2.36 | 30.0–3.15 | 50.0–3.0 | 30.0-1.55 | |
| No. of reflections | 57,871 | 51,122 | 54,107 | 57,455 | |
| Rwork/Rfree (%) | 20.8/24.6 | 22.3/25.6 | 21.1/24.5 | 23.2/26.7 | |
| Protein atoms | 8,298 | 16,536 | 16,536 | 3,933 | |
| DNA atoms | 290 | 674 | 0 | 0 | |
| Cofactor atoms | 41 | 70 | 70 | 0 | |
| Rmsd bond lengths (Å) | 0.009 | 0.009 | 0.009 | 0.007 | |
| Rmsd bond angles (°) B factors (Å2): protein DNA Ca2+ water Wilson | 1.4 65.5 107.6 58.9 49.5 58.4 | 1.4 | 1.46 | 1.3 |
-
*Only the ssDNA is ordered. Values in parentheses are for the highest-resolution shell.





