Deficiency in DNAH12 causes male infertility by impairing DNAH1 and DNALI1 recruitment in humans and mice
Figures
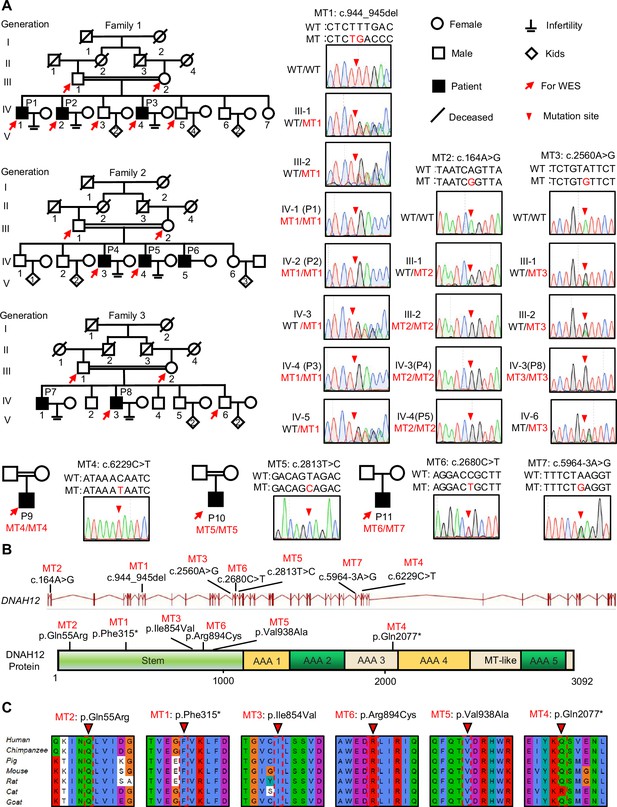
Identification of bi-allelic DNAH12 variants in infertile men with asthenoteratozoospermia.
(A) Pedigrees of family 1 with three infertile males, P1 (IV:1), P2 (IV:2), and P3 (IV:4), family 2 with three infertile males, P4 (IV:3), P5 (IV:4) and P6 (IV:5), family 3 with two infertile males, P7 (IV:1) and P8 (IV:3), and three sporadic infertile cases P9, P10 and P11. Red arrows point to the individuals for whom whole-exome sequencing was performed. Candidate mutations were validated by Sanger sequencing and the chromatograms were shown on the right. Red arrowheads point to mutant sites. Double horizontal lines represent consanguineous marriages. WT, wild-type allele; MT, the mutant allele. (B) The positions of the DNAH12 variants at the transcript (ENST00000351747.2) and protein levels (Q6ZR08-1). MT-like, Microtubule-binding-like domain. (C) Conservation analyses of affected amino acids across different species, the arrowheads indicate the mutation sites.
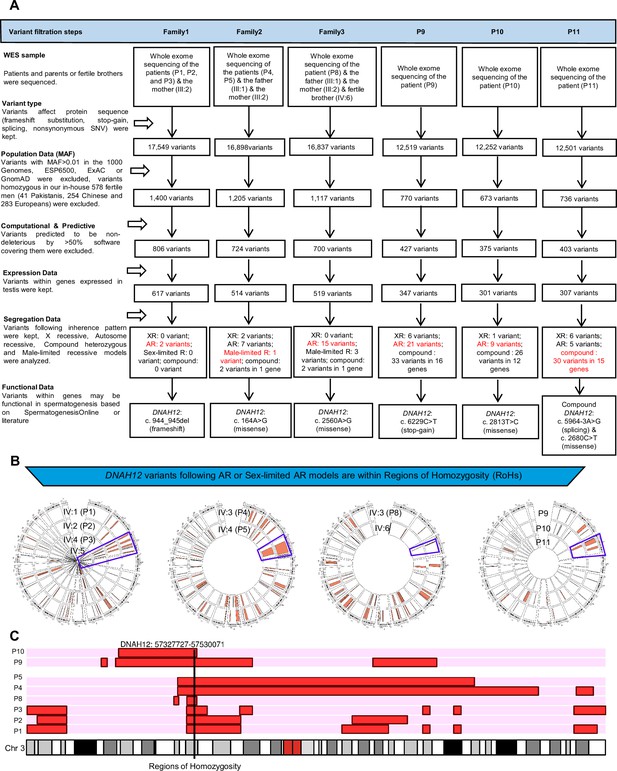
Flowchart for analyses of the whole-exome sequencing data.
(A) In-silico analyses and filtering of variants in patients from Pakistani and Chinese families. (B) and (C) DNAH12 variants are located in the regions of homozygosity (RoHs) of affected Pakistani or Chinese patients. RoHs are marked in red. The purple frames mark the location of the mutations.
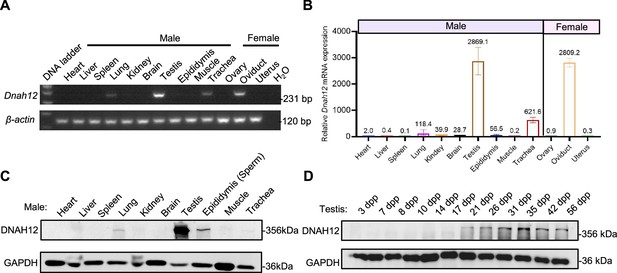
Expression pattern of Dnah12 in different mouse tissues and developmental stages of the testis.
(A) RT‒PCR analysis of Dnah12 expression in different tissues from adult mice. β-actin served as the reference gene. (B) Real-time quantitative PCR for Dnah12 transcripts in various 8-wk-old mouse tissues (most organs were obtained from male mice, except ovary, oviduct, and uterus which were gained from female mice), β-actin gene served as reference, and the experiments were repeated three times. (C) Immunoblotting assay of the DNAH12 expression among 10 different tissues from adult mice. (D) Immunoblotting analysis of the testicular DNAH12 expression at different days postpartum (dpp). GAPDH served as the loading control.
-
Figure 1—figure supplement 2—source data 1
Labelled files for western blot in Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig1-figsupp2-data1-v1.zip
-
Figure 1—figure supplement 2—source data 2
Original files for western blot in Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig1-figsupp2-data2-v1.zip
-
Figure 1—figure supplement 2—source data 3
Labelled file for gel in Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig1-figsupp2-data3-v1.zip
-
Figure 1—figure supplement 2—source data 4
Original file for gel in Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig1-figsupp2-data4-v1.zip

Phylogenetic analysis of the DNAH12 homologous proteins in different species.
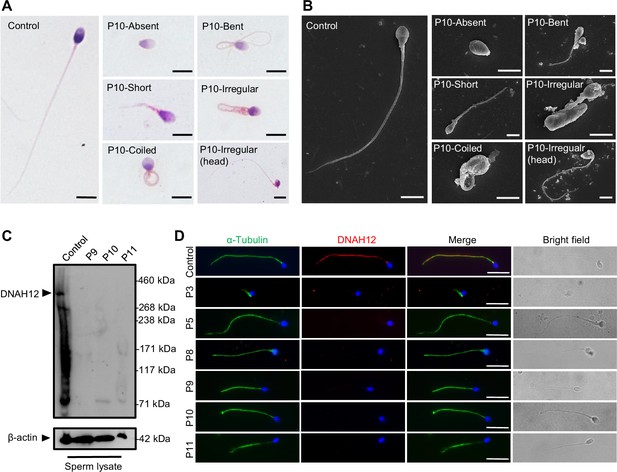
Abnormal sperm morphology and undetectable DNAH12 expression in men with bi-allelic DNAH12 variants.
(A) Representative micrographs show normal sperm morphology from a fertile control and abnormal spermatozoa from P10 with absent, short, coiled, bent, irregular flagella, or abnormal head under light microscopy. Scale bars, 5 μm. (B) Representative scanning electron microscopy (SEM) micrographs show details about the sperm morphology of a fertile control and P10. The normal sperm morphology can be observed in the fertile control (left) and abnormal types like absent, short, coiled, bent, irregular tails, and head abnormality in P10 (right). Scale bars, 5 μm. (C) Immunoblotting assay revealed that DNAH12 was absent in the spermatozoa from P9, P10, and P11 harboring DNAH12 variants. β-actin was used as a loading control. (D) Sperm cells were co-stained with α-Tubulin (green) and DNAH12 (red) antibodies while no DNAH12 signals were observed in sperm flagella of patients. DNA was counterstained with Hoechst 33342. Scale bars, 10 μm.
-
Figure 2—source data 1
Labelled file for western blot in Figure 2A.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig2-data1-v1.zip
-
Figure 2—source data 2
Original file for western blot in Figure 2A.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig2-data2-v1.zip
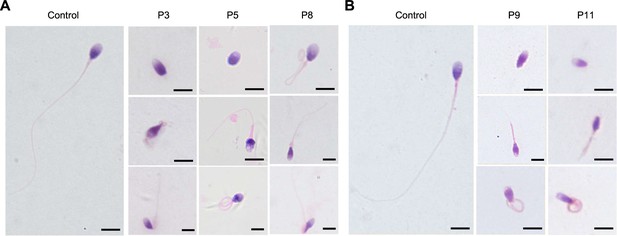
Sperm morphology of fertile controls and patients.
(A) Representative micro-graphs from men harboring bi-allelic DNAH12 variants (P3 of family 1, P5 of family 2, and P8 of family 3) and a fertile control by H&E staining. (B) Representative micrographs from P9, P11, and a fertile control by Papanicolaou staining. Scale bars,10 µm.
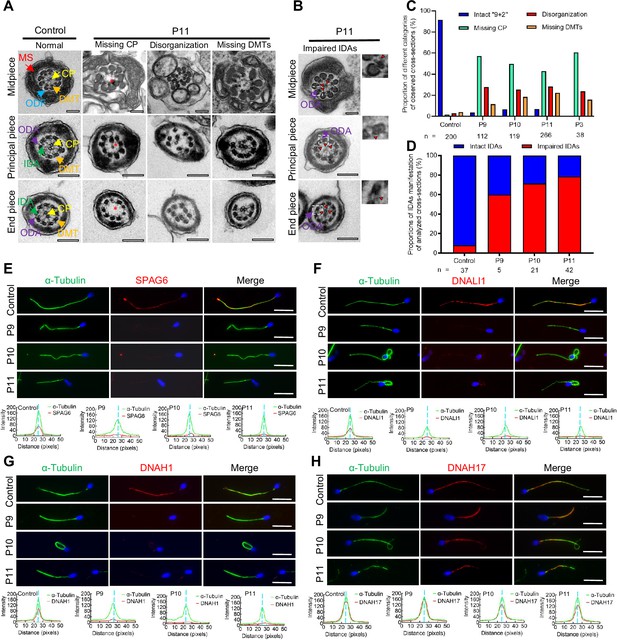
Flagellar axoneme defects were detected in the patients with DNAH12 variants.
(A–B) Representative transmission electron microscopy (TEM) micrographs showing cross-sections of the midpiece, principal piece, and end piece of sperm flagella from a fertile control and P11. The axoneme structure in control presents a ‘9+2’ microtubules arrangement, including mitochondrial sheath (MS, indicated with red arrows), central pair of microtubules (CP, indicated with yellow arrows), outer dense fibers (ODFs, indicated with cerulean arrows), peripheral doublet microtubules (DMTs, indicated with orange arrows), inner dynein arms (IDAs, indicated with green arrows), and outer dynein arms (ODAs, indicated with purple arrows), while in P11, axonemal defects like missing CP, disorganized axonemal structures or missing DMTs were observed, the red asterisks mark CP loss (A). Impaired IDAs in the sperm axoneme of patient P11 (B). The red triangle marks impaired IDAs while adjacent ODAs are identifiable. Scale bars, 200 nm. (C–D) The proportion of different categories of observed cross-sections in the control and patients. Cross-sections were classified into four categories: Intact ‘9+2’, missing CP, disorganization, and missing DMTs (C), and the proportion of intact IDAs or impaired IDAs in the cross-sections of flagellar axoneme of which ODAs were identifiable (D). n, the total number of cross-sections for quantification. (E–G) Representative images of spermatozoa from fertile controls and patients carrying bi-allelic DNAH12 variants co-stained α-Tubulin with SPAG6 (E), DNAH1 (F), DNALI1 (G), or DNAH17 (H), and Hoechst 33342 for DNA (blue). The fluorescent signal intensity profiles were shown on the bottom. Scale bars, 10 µm.
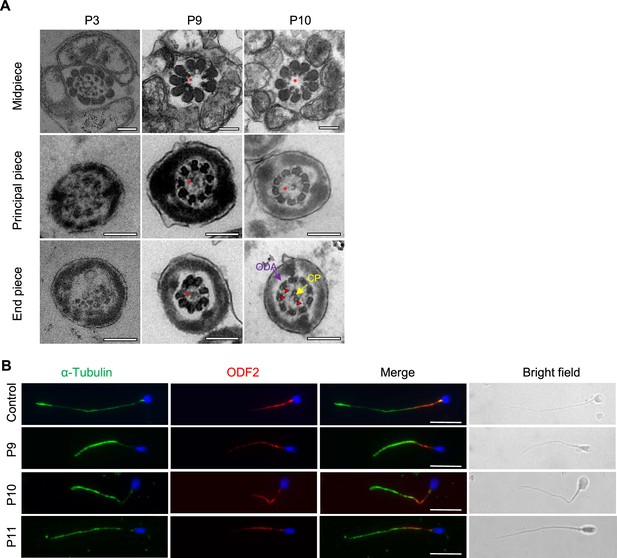
Transmission electron microscopy (TEM) images of men harboring bi-allelic DNAH12 variants and immuno-fluorescence assay of ODF2 in sperm of control, P9, P10 and P11.
(A) Representative TEM micrographs from men harboring bi-allelic DNAH12 variants (P3 of Famliy1, P9 and P10). Red asterisks mark central pair (CP) loss and red triangle marks the positions where inner dynein arms (IDAs) are impaired or absent while adjacent outer dynein arms (ODAs) are identifiable. Scale bars, 200 nm. (B) Representative image of spermatozoa from fertile controls and patients carrying bi-allelic DNAH12 variants stained with ODF2 and α-Tubulin antibodies, and Hoechst 33342 (blue). Scale bars,10 µm.
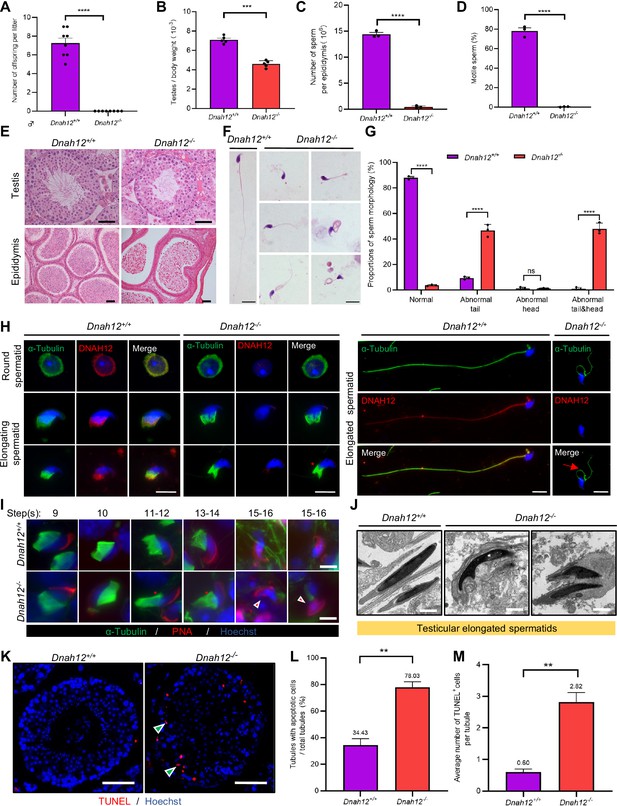
Disruption of Dnah12 results in impaired spermatogenesis and abnormal spermatozoa morphology.
(A) Fertility test of adult Dnah12+/+ and Dnah12-/- male mice (n=8 independent experiments). Data are presented as mean ± SEM; ****p<0.0001. (B) The testes to body weight ratios of Dnah12+/+ and Dnah12-/- mice (n=5 independent experiments). Data are presented as mean ± SEM; ***p<0.001. (C) The number of sperm per epididymis of Dnah12+/+ and Dnah12-/- male mice (n=3 independent experiments). Data are presented as mean ± SEM; ****p<0.0001. (D) Percentages of motile spermatozoa in Dnah12+/+ and Dnah12-/- male mice (n=3 independent experiments). Data are presented as mean ± SEM; ****p<0.0001. (E) Histological sections of testis and epididymis from Dnah12+/+ and Dnah12-/- mice after H&E staining. Scale bars, 50 μm. (F) Morphology of the sperm from caudal epididymis of Dnah12+/+ and Dnah12-/- mice. Scale bars,10 μm. (G) Quantitative analysis of sperm morphology of Dnah12+/+ and Dnah12-/- male mice. The experiments were repeated three times with at least 200 spermatozoa counted every time. The above data were obtained from three adult mice for each genotype (n=3 independent experiments). Data are presented as mean ± SEM; ns indicates no significant difference; ****p<0.0001. (H) Representative images of testicular cells from Dnah12+/+ and Dnah12-/- mice co-stained DNAH12 and α-Tubulin antibodies. Scale bars, 10 μm. The red asterisk indicates abnormal manchette structure and the red arrow indicates abnormal sperm flagellum at the elongated spermatid stage of Dnah12-/- mice. (I) Representative immunofluorescence images of seminiferous tubule squash from 8-wk-old Dnah12+/+ and Dnah12-/- mice, co-stained by peanut agglutinin (PNA) (red) and α-Tubulin (green) antibody. DNA was stained with Hoechst 33342. Scale bars, 10 μm. The asterisks indicate the manchette defects and the red triangles mark the abnormal sperm heads. Scale bars,10 μm. (J) Representative transmission electron microscopy (TEM) micrographs of testicular elongated spermatids from Dnah12+/+ and Dnah12-/- mice. Scale bars, 1 μm. (K) TUNEL assay on the testicular sections from Dnah12+/+ and Dnah12-/- mice. The green triangles mark the apoptotic elongating or elongated spermatids. Scale bars, 50 μm. (L) The proportion of tubules with apoptotic cells in Dnah12+/+ and Dnah12-/- mice. (M) Average numbers of TUNEL-positive cells per tubule in Dnah12+/+ and Dnah12-/- mice (n=3 independent experiments). Data are obtained from three mice with at least 50 tubules scored in each repeated experiment. Data are presented as mean ± SEM; **p<0.01.
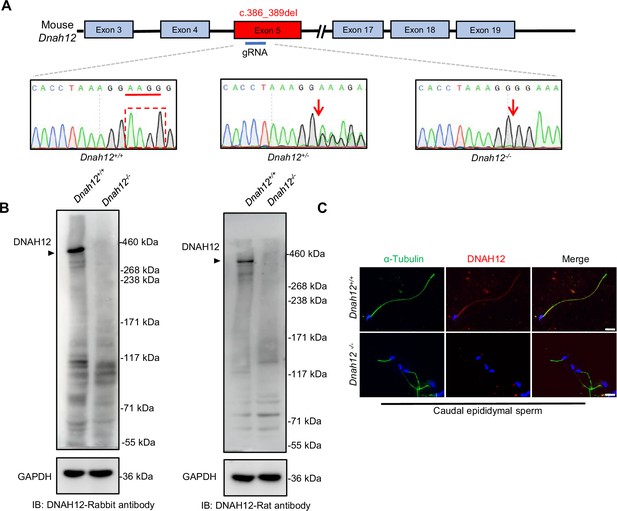
Generation of Dnah12-/- mice model and validation of rabbit and rat host DNAH12 antibodies.
(A) Schematic illustrating construction of the Dnah12−/− mouse model and Sanger sequencing validation of Dnah12+/+, Dnah12+/-, and Dnah12-/- mice. Red arrows indicate the position where 4 bp deletion occurs. (B) Immunoblotting assay of testis lysate from 10 wk Dnah12+/+ and Dnah12-/- mice using DNAH12-Rabbit antibody or DNAH12-Rat antibody. (C) Representative images of caudal epididymal sperm from Dnah12+/+ and Dnah12-/- mice co-stained α-Tubulin and DNAH12 antibodies. Scale bars, 10 μm.
-
Figure 4—figure supplement 1—source data 1
Labelled files for western blot in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original files for western blot in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig4-figsupp1-data2-v1.zip
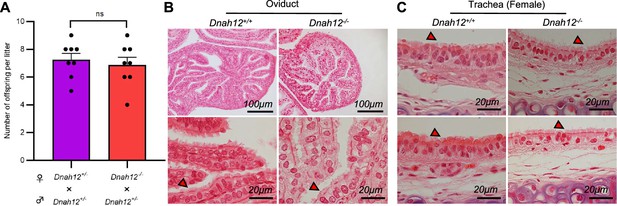
Normal development and fertility in Dnah12-/- female mice.
(A) Number of offspring per litter of Dnah12+/- and Dnah12-/- from 8–10 wk old female mice, the two groups were caged with Dnah12+/- mice, and the number of offspring was recorded, respectively (n=8 independent experiments).Data are presented as mean ± SEM; ns indicates no significant difference. (B–C) Oviductal (B) or tracheal (C) histology of Dnah12+/+. The red triangles mark the cilia with normal morphology in Dnah12-/- 8-wk-old female mice and the cilia shapes and lengths were similar in the two groups. Scare bars are shown in the figures.
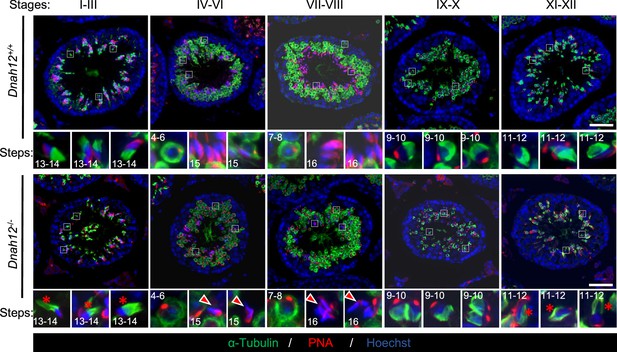
Deficiency in manchette organization and abnormal sperm head shaping during spermiogenesis in Dnah12-/- mice.
Representative images of seminiferous tubule sections from 8-wk-old Dnah12+/+ and Dnah12-/- mice, co-stained by peanut agglutinin (PNA) (red) and α-Tubulin (green) antibodies. DNA was stained with Hoechst 33342. Scale bars, 50 μm.
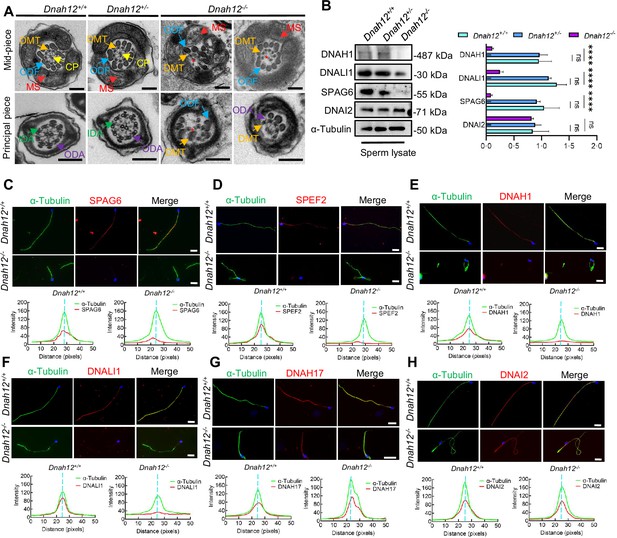
Deletion of Dnah12 causes axoneme defects in mice.
(A) Representative transmission electron microscopy (TEM) micrographs showing cross-sections of sperm flagella from Dnah12+/+, Dnah12+/-, and Dnah12-/- mice. In Dnah12+/+, Dnah12+/-, the normal axoneme ‘9+2’ microtubule arrangement mainly consisted of the mass spectrometry (MS), (indicated with red arrows), central pair (CP) (yellow arrows), outer dense fibers (ODFs) (cerulean arrows), doublet microtubules (DMTs) (orange arrows), inner dynein arms (IDAs) (green arrows), and outer dynein arms (ODAs) (purple arrows). However, the CP and DMTs structures were missing or disarranged in the Dnah12-/- group. Red asterisks mark the loss of CP. Scale bars, 200 nm. (B) Immunoblotting of sperm lysate from Dnah12+/+, DNAH12+/- and Dnah12-/- mice using DNAH1, DNALI1 or SPAG6 antibodies. DNAI2 and α-Tubulin were used as the loading controls. The relative band grayscales of proteins to α-Tubulin in sperm lysate from Dnah12+/+, DNAH12+/-, and Dnah12-/- mice were shown on the right (n=4 independent experiments). Data are presented as mean ± SEM; ns indicates no significant difference; ****p<0.0001. (C–H) Representative images of caudal epididymal sperm from Dnah12+/- and Dnah12-/- mice co-stained α-Tubulin and SPAG6 (C), SPEF2 (a component of CP complex) (D), DNAH1 (E), DNALI1 (F), DNAH17 (G) or DNAI2 (H) antibodies. The fluorescent signal intensity profiles were shown on the bottom. Scale bars, 10 μm.
-
Figure 5—source data 1
Labelled files for western blot in Figure 5B.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig5-data1-v1.zip
-
Figure 5—source data 2
Labelled files for western blot in Figure 5B.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig5-data2-v1.zip
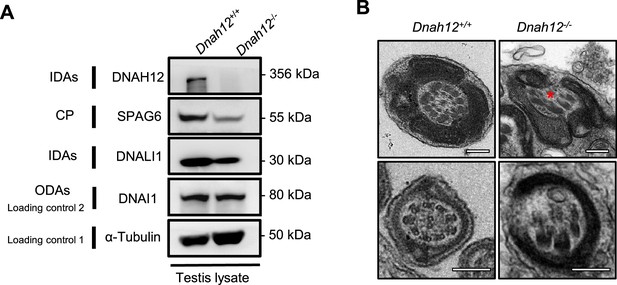
The central pair (CP) and doublet microtubules (DMTs) structures were impaired in the Dnah12-/- testicular sperm axoneme.
(A) Immunoblotting of testis lysate from Dnah12+/+ and Dnah12-/- mice using SPAG6, DNALI1 antibodies. DNAI1 and α-Tubulin were used as the loading controls. (B) Representative TEM micrographs showing cross-sections of testicular sperm axoneme from Dnah12+/+ and Dnah12-/- mice. The CP and DMTs were missing or disarranged in the Dnah12-/- group. Red asterisks mark the CP loss. Scale bars, 200 nm.
-
Figure 5—figure supplement 1—source data 1
Labelled files for western blot in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Original files for western blot in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig5-figsupp1-data2-v1.zip
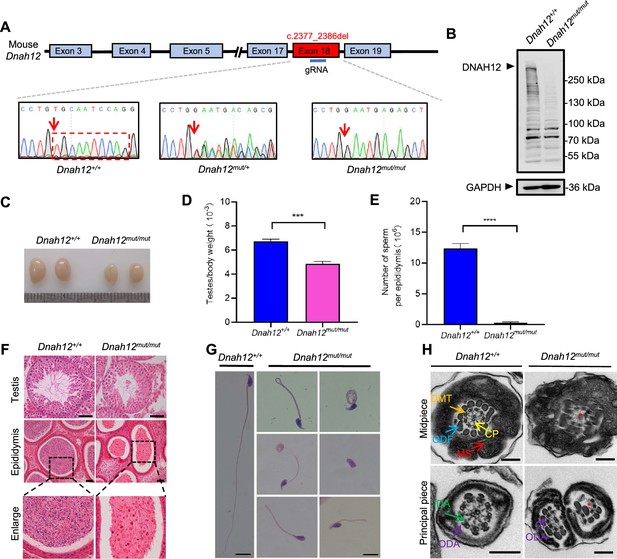
Generation of Dnah12mut/mut mice.
(A) Schematic illustrating construction of the Dnah12mut/mut mice model and Sanger sequencing results of Dnah12+/+, Dnah12+/mut, and Dnah12mut/mut mice. The red arrows indicate the position where the mutation occurs. (B) Immunoblotting of testicular lysate from Dnah12+/+ and Dnah12mut/mut mice using homemade DNAH12-Rabbit antibody. GAPDH was used as a loading control. The arrowheads indicate the target bands. (C) Representative images of testes from Dnah12+/+ and Dnah12mut/mut mice. (D) Testes to body weight ratios of Dnah12+/+ or Dnah12mut/mut mice (n=3 independent experiments). Data are presented as mean ± SEM; ***p<0.001. (E) Number of sperm per epididymis of Dnah12+/+ and Dnah12mut/mut mice (n=3 independent experiments). Data are presented as mean ± SEM; ****p<0.0001. (F) Histological sections of testis and epididymis from Dnah12+/+ and Dnah12mut/mut after H&E staining. Scale bars, 50 μm. (G) Morphology of the spermatozoa obtained from Dnah12+/+ mice or Dnah12mut/mut mice cauda epididymis after H&E staining. Scale bars,10 μm. (H) Representative transmission electron microscopy (TEM) micrographs showing cross sections of sperm flagella from Dnah12+/+ and Dnah12mut/mut mice. Scale bars, 200 nm.
-
Figure 5—figure supplement 2—source data 1
Labelled files for western blot in Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig5-figsupp2-data1-v1.zip
-
Figure 5—figure supplement 2—source data 2
Original files for western blot in Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig5-figsupp2-data2-v1.zip
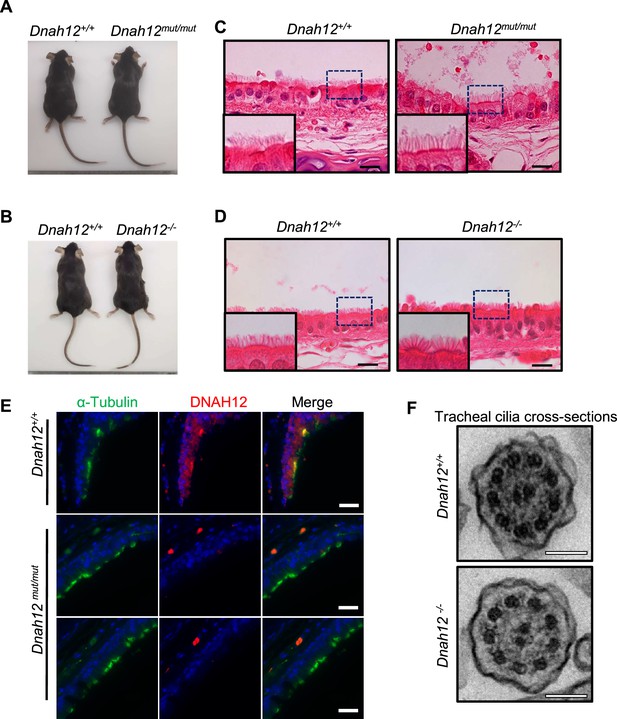
Normal development and no obvious primary ciliary dyskinesia (PCD) symptoms were observed in Dnah12mut/mut and Dnah12-/- males.
(A–B) No obvious developmental abnormalities were observed in adult Dnah12mut/mut (A) or Dnah12-/- (B) mice. (C–D) Tracheal histology showed the cilia shape and lengths of Dnah12mut/mut (C) or Dnah12-/- (D) were similar to Dnah12+/+ in the two groups. Scare bars, 20 μm. (E) Representative images of tracheal cilia co-stained α-Tubulin (green) and DNAH12 (red) antibodies. Scale bars, 20 μm. (F) TEM micrographs showing cross sections of tracheal cilia cross-sections from Dnah12+/+ and Dnah12-/- mice. Scale bars, 200 nm.
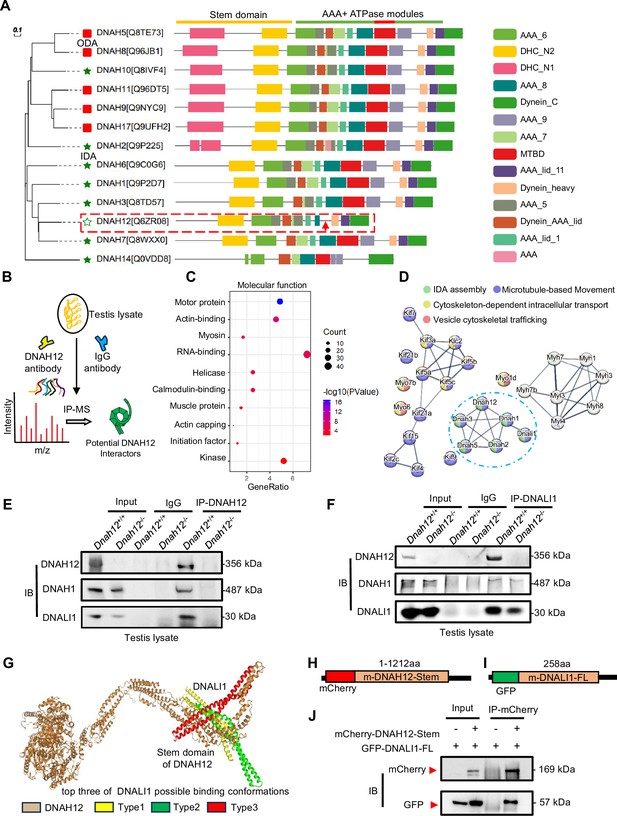
DNAH12 interacts with inner dynein arm (IDA) components DNALI1 and DNAH1.
(A) Phylogenetic and protein domain analyses of the DNAH family. The dashed box indicates the shortest-length member, DNAH12, while the arrow points to the missing microtubule-binding domain (MTBD) region. (B) Schematic diagram showing the procedure of the immunoprecipitation/mass spectrometry (IP/MS) proteomics experiment. (C) GO term enrichment analysis of DNAH12 potential interactors. (D) Protein-protein interaction network of DNAH12 among the motor protein term. The blue circle marks the proteins involved in IDA assembly. (E–F) Co-immunoprecipitation (Co-IP) assays using DNAH12 antibody (E) or DNALI1 antibody (F) showed strong and specific interactions among DNAH12, DNALI1, and DNAH1. (G) Predicted DNAH12-DNALI1 interaction details by the HDOCK server. The top three of DNALI1 possible binding conformations were shown in types 1–3. (H–J) Construction of mouse-mCherry-DNAH12-Stem (H) and mouse-GFP-DNALI1-full length (FL) (I) plasmids, and the validation of the interaction between the Stem domain of DNAH12 and full-length of DNALI1 (J).
-
Figure 6—source data 1
Labelled files for western blot in Figure 6E, F and J.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig6-data1-v1.zip
-
Figure 6—source data 2
Original files for western blot in Figure 6E, F and J.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig6-data2-v1.zip
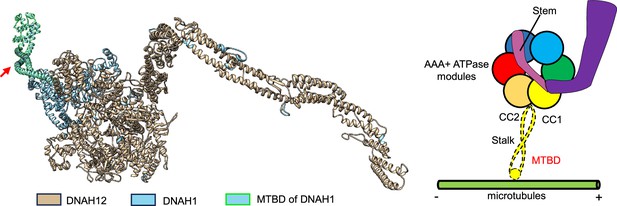
DNAH12 lacks the microtubule-binding domain (MTBD) domain.
The three-dimensional protein models of human DNAH12, DNAH1, and DNAH12 were colored golden while DNAH1 was colored celeste, for better visualization, the MTBD of DNAH1 was box selected with green lines. The proposed cartoon structures of the DNAH family are shown on the right, while DNAH12 lacks the MTBD domain.
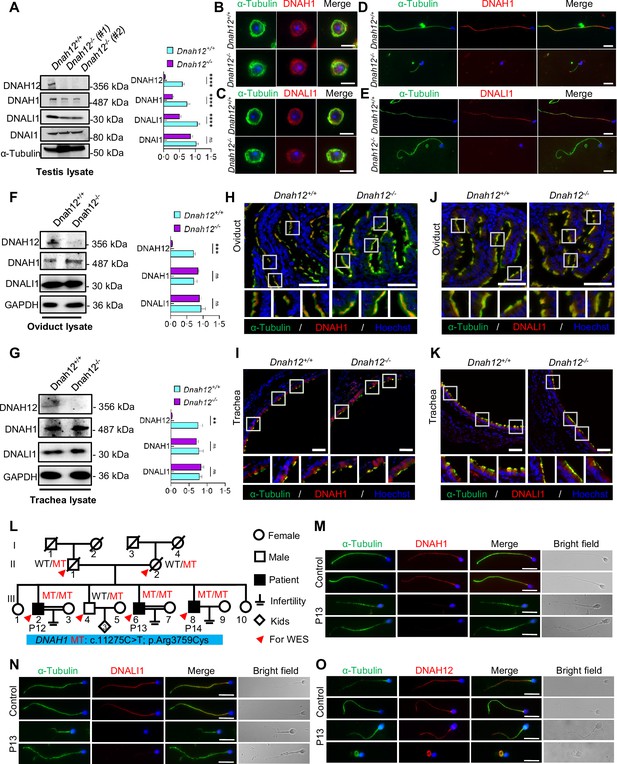
DNAH12 facilitates the recruitment of DNALI1 and DNAH1 to flagella, but not cilia.
(A) Immunoblotting of testis lysate from Dnah12+/+, DNAH12+/- and Dnah12-/- mice using DNAH12, DNAH1, and DNALI1 antibodies. DNAI1 (a component of ODAs) and α-Tubulin were used as the loading controls. The relative band grayscales of proteins to α-Tubulin in testis lysate from Dnah12+/+, and Dnah12-/- mice were shown on the right (n=5 independent experiments). Data are presented as mean ± SEM; ns indicates no significant difference;****p<0.0001. (B–C) Immunofluorescence assays of α-Tubulin and DNAH1 (B), or DNALI1 (C) antibodies on round spermatids from adult Dnah12+/− and Dnah12−/− mice. Scale bars, 10 μm. (D–E) Immunofluorescence assays of α-Tubulin and DNAH1 (D), or DNALI1 (E) antibodies on elongated spermatids from adult Dnah12+/− and Dnah12−/− mice. Scale bars, 10 μm. (F–G) Immunoblotting of oviduct (F) or trachea (G) lysate from Dnah12+/+, DNAH12+/- and Dnah12-/- mice using DNAH12, DNAH1, and DNALI1 antibodies. GAPDH was used as the loading control. The relative band grayscales of proteins to GAPDH in oviduct lysate (F) or trachea lysate (G) from Dnah12+/+, and Dnah12-/- mice were shown on the right (n=4 independent experiments). Data are presented as mean ± SEM; ns indicates no significant difference; ***p<0.001. (H–I) Immunofluorescence assays of the oviduct (H) or trachea (I) sections with α-Tubulin and DNAH1 antibodies. Scale bars, 50 μm. (J–K) Immunofluorescence assays of the oviduct (J) or trachea (K) sections with α-Tubulin and DNALI1 antibodies. Scale bars, 50 μm. (L) Pedigree of a family with three infertile males, P12 (III-2), P13 (III:6), and P14 (III:8); MT: the DNAH1 mutation c.11275C>T; p.Arg3759Cys. (M–O) Representative images of spermatozoa from a fertile control and P13 co-stained by α-Tubulin antibody and DNAH1 (M), DNALI1(N), or DNAH12 (O) antibodies, respectively. Scale bars,10 μm.
-
Figure 7—source data 1
Labelled files for western blot in Figure 7.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig7-data1-v1.zip
-
Figure 7—source data 2
Original files for western blot in Figure 7.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig7-data2-v1.zip
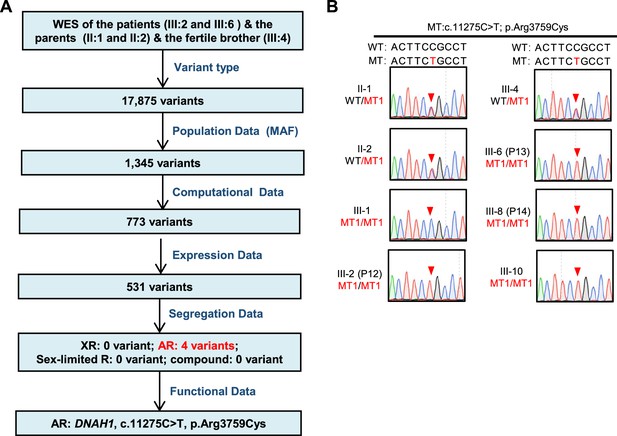
Flowchart for analyses of the whole-exome sequencing of a Pakistani family and validation of DNAH1 mutation through Sanger sequencing.
(A) Flowchart for analyses of the whole-exome sequencing of a Pakistani family with three infertile male patients. (B) Validation of the candidate homozygous DNAH1 mutation c.11275C>T, p.Arg3759Cys in the family members by Sanger sequencing.
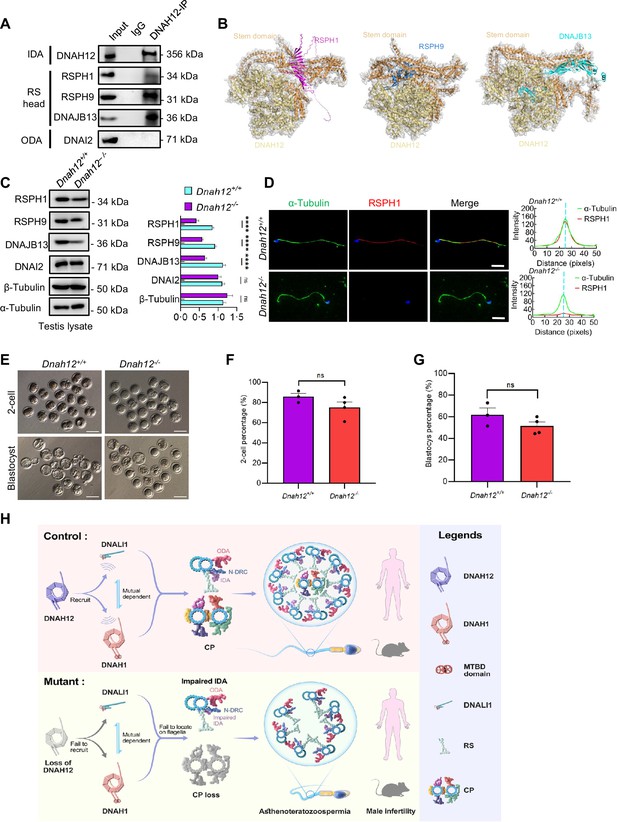
DNAH12 may interact with RS head-associated proteins to regulate CP stability.
(A) Co-immunoprecipitation (Co-IP) assays of the potential interactions of DNAH12 and RS head proteins RSPH1, RPSH9, or DNAJB13. DNAI2 antibody was used as a negative control. (B) Interaction analyses of DNAH12 and RS head proteins RSPH1, RPSH9, or DNAJB13 by AlphaFold3. (C) Immunoblotting of testis lysate from Dnah12+/+ and Dnah12-/- mice using RSPH1, RPSH9, DNAJB13 antibodies. DNAI2 (a component of ODAs), α-Tubulin, and β-Tubulin were used as the loading controls. The relative band grayscales of proteins to α-Tubulin in testis lysate from Dnah12+/+, and Dnah12-/- mice were shown on the right (n=4 independent experiments). Data are presented as mean ± SEM; ns indicates no significant difference;****p<0.0001.; (D) Immunofluorescence assays of α-Tubulin and RSPH1 antibodies on sperm collected from Dnah12+/− and Dnah12−/− mice caudal epididymis. The fluorescent signal intensity profiles were shown on the right. Scale bars, 10 μm. (E) Representative two-cell embryos and blastocysts of Dnah12+/+, and Dnah12-/ male mice after intracytoplasmic sperm injection. Scale bar, 100 μm.(F) Percentages of two-cell-stage embryos of Dnah12+/+(n=3 mice), and Dnah12-/- male mice (n=4 mice) after intracytoplasmic sperm injection. Data are presented as mean ± SEM; ns indicates no significant difference. (G) Percentages of blastocyst-stage embryos of Dnah12+/+(n=3 mice), and Dnah12-/- males (n=4 mice) after intracytoplasmic sperm injection. Data are presented as mean ± SEM; ns indicates no significant difference. (H) Schematic diagram showing the proposed function of DNAH12 in sperm flagellar development in humans and mice. DNAH12 is essential for recruiting DNALI1 and DNAH1 to sperm flagella and maintaining the proper axonemal arrangement, especially IDAs and CP structures. Loss of DNAH12 causes the failure of DNALI1 and DNAH1 to be recruited to sperm flagella and results in abnormal sperm morphology with compromised axoneme organization, causing male infertility. ODA, outer dynein arm; IDA, inner dynein arm; N-DRC, nexin-dynein regulatory complex; CP, central pair of microtubules; MTBD, microtubule-binding domain; RS, radial spoke.
-
Figure 8—source data 1
Labelled files for western blot in Figure 8A and C.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig8-data1-v1.zip
-
Figure 8—source data 2
Original files for western blot in Figure 8A and C.
- https://cdn.elifesciences.org/articles/100350/elife-100350-fig8-data2-v1.zip
Videos
Sperm motility of caudal sperm from Dnah12+/+ mice.
Sperm motility of caudal sperm from Dnah12-/- mice.
Tracheal ciliary motility of Dnah12+/+ mice.
Tracheal ciliary motility of Dnah12-/- mice.
Tables
Detailed information of bi-allelic DNAH12 variants identified in infertile men.
The accession number of human DNAH12 is ENST00000351747.2. (b) Abbreviations are listed as follows: SNV, single nucleotide variants; NA, not assessed; T, tolerated; D, deleterious. SIFT, Sorting Intolerant From Tolerant; PolyPhen-2, Polymorphism Phenotype v2; FATHMM-MKL, Functional Analysis Through Hidden Markov Models-Multiple Kernel Learning; CADD, Combined Annotation-Dependent Depletion; and a mutation is predicted deleterious if the CADD Phred score >20 in CADD. gnomAD, Genome Aggregation Database.
| Genomic position on chr3 | cDNAChange a | Mutation type | Genetic pattern | SIFT | Polyphen2 | fathmm-MKL | CADD | 1000 Genomes | gnomAD | |
|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 | 57,489,883 | MT1: c.944_945del | frameshift substitution | AR | NA | NA | NA | NA | 0 | 0.00002844 |
| Family 2 | 57,528,434 | MT2: c.164A>G | nonsynonymous SNV b | Male-limited AR | T | D | D | D | 0.000599042 | 0.0004 |
| Family 3 | 57,447,323 | MT3: c.2560A>G | nonsynonymous SNV | AR | T | D | D | D | 0.000599042 | 0.0002 |
| P9 | 57,391,670 | MT4: c.6229C>T | stop-gain | AR | NA | NA | D | NA | 0 | 0.00002714 |
| P10 | 57,445,368 | MT5: c.2813T>C | nonsynonymous SNV | AR | D | D | D | D | 0.000199681 | 0.00003383 |
| P11 | 57,394,265 | MT6: c. 5964–3A>G | splicing | AR | NA | NA | NA | NA | 0.000199681 | 0.0002 |
| 57,445,501 | MT7: c.2680C>T | nonsynonymous SNV | D | D | D | D | 0 | 0.000007178 |
Clinical characteristics of the infertile patients with bi-allelic DNAH12 mutations.
The reference limits are shown according to WHO sixth edition standards. (b) Reference values are shown by observation of sperm morphology in three fertile individuals. (c) ‘Age’ represents age in 2024. ND, not determined.
| Reference values a | Fertile controls b | Family1 | Family2 | Family3 | P9 | P10 | P11 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IV-1(P1) | IV-2(P2) | IV-4(P3) | IV-5(P4) | IV-7(P5) | IV-4(P8) | ||||||
| cDNA mutation | – | – | MT1/MT1 | MT1/MT1 | MT1/MT1 | MT2/MT2 | MT2/MT2 | MT3/MT3 | MT4/MT4 | MT5/MT5 | MT6/MT7 |
| Age (y) c | – | – | 58 | 49 | 42 | 40 | 38 | 32 | 33 | 40 | 31 |
| Semen parameters | |||||||||||
| Semen volume (ml) | >1.4 | – | ND | 1.5±0.5 | 2.8±0.3 | 4.0 | 3.0 | 3.0 | 3.2 | 3.1 | 2.4 |
| Sperm concentration (106 /ml) | >15 | – | ND | 58±28.5 | 50.0±15.0 | 6.0 | 8.0 | 2.8 | 24.7 | 83.1 | 57.1 |
| Semen pH | Alkaline | – | ND | Alkaline | Alkaline | Alkaline | Alkaline | Alkaline | Alkaline | Alkaline | Alkaline |
| Motile sperm (%) | >40 | – | ND | 29.3±9.0 | 7.5±7.5 | 14.0 | 20.0 | 20 | 15.1 | 17.4 | 7.4 |
| Progressively motile sperm (%) | >30 | – | ND | 13.3±10.3 | 16.7±17 | 5.0 | 8.0 | 5 | 13.4 | 16.8 | 24 |
| Sperm morphology | |||||||||||
| Normal (%) | – | 76.5±2.0 | ND | ND | 19.9 | ND | 21.7 | 17.0 | 14.0 | 17.4 | 7.4 |
| Abnormal head (%) | – | 7.0±1.9 | ND | ND | 16.4 | ND | 5.4 | 6.0 | 3.5 | 16.4 | 20.0 |
| Abnormal tail (%) | – | 12.4±3.4 | ND | ND | 43.8 | ND | 41.4 | 44.0 | 42.4 | 37.4 | 16.3 |
| Abnormal head &tail (%) | – | 4.1±1.3 | ND | ND | 19.9 | ND | 31.5 | 33.0 | 40.2 | 28.8 | 56.3 |
| Sperm flagella morphology | |||||||||||
| Normal (%) | – | 83.1±2.3 | ND | ND | 26.4 | ND | 26.6 | 22.7 | 17.9 | 31.7 | 27.4 |
| Absent (%) | – | 2.7±1.1 | ND | ND | 16.4 | ND | 7.9 | 15.8 | 9.6 | 16.6 | 11.6 |
| Short (%) | – | 3.6±0.6 | ND | ND | 16.4 | ND | 11.3 | 14.9 | 20.1 | 11.7 | 8.4 |
| Coiled (%) | – | 2.8±0.7 | ND | ND | 17.3 | ND | 24.6 | 12.9 | 11.8 | 14.6 | 7.0 |
| Bent (%) | – | 6.3±0.6 | ND | ND | 12.7 | ND | 15.8 | 10.0 | 13.5 | 7.8 | 33.5 |
| Irregular caliber (%) | – | 1.5±0.2 | ND | ND | 10.9 | ND | 13.8 | 23.8 | 27.1 | 17.6 | 12.1 |
Additional files
-
Supplementary file 1
The used reagents and protein interactors information.
(a) List of candidate dynein protein interactors of DNAH12. (b) Primers used in the study. Primer used for Sanger Sequencing of the identified mutations, genotyping of the mice models, Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Quantitative Polymerase Chain Reaction (qPCR). (c) The primary antibodies used in this study. (d) The secondary antibodies used in this study. (e) Primers used for the construction of plasmids in co-immunoprecipitation (Co-IP assays).
- https://cdn.elifesciences.org/articles/100350/elife-100350-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/100350/elife-100350-mdarchecklist1-v1.docx





