The Drosophila hematopoietic niche assembles through collective cell migration controlled by neighbor tissues and Slit-Robo signaling
Figures
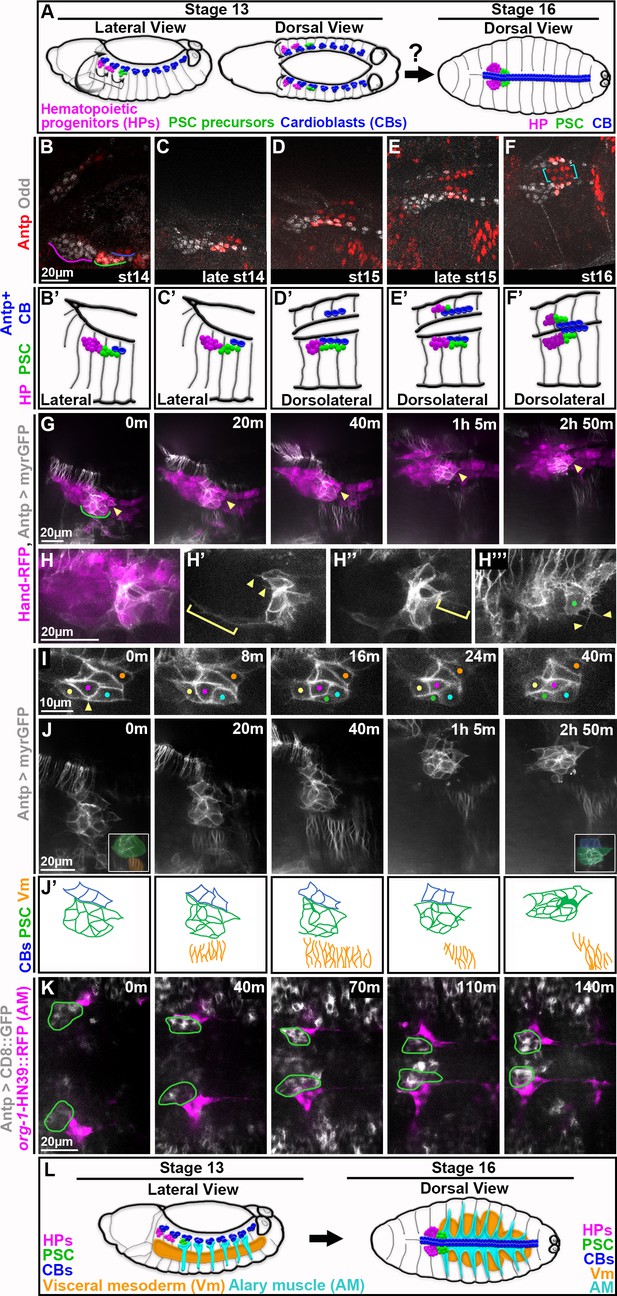
Live-imaging reveals dynamics of PSC migration and presence of nearby muscles.
All images are oriented with anterior to the left and posterior to the right. (A) Schematic depicts prior knowledge of PSC migration. Prospective cells of lymph glands (hematopoietic progenitors, magenta, and PSC, green) and cardioblasts (blue) of the dorsal vessel are specified as distinct clusters prior to migration onset around stage 13 (left). Around stage 16 (right) lymph gland clusters have coalesced and flank the dorsal vessel at the dorsal midline; bilaterally symmetric counterparts are aligned. (B-F) Fixed w1118 embryos stained for Antp and Odd to label the PSC. (B) Stage 14 PSC (green) is lateral in embryo, tightly clustered, and flanks 2 Antp + CBs (blue). (C) Late stage 14 PSC is more dorsal, flanking 2 Antp + CBs. (D) The stage 15 PSC is even more dorsal, elongated, and flanks 4 Antp + CBs; contralateral Antp + CBs in view. (E) Late stage 15 PSC is more medial and compact. (F) Stage 16 LGs and CBs at final position at dorsal midline, right LG partially in view. PSC is compact and coalesced at LG posterior, flanking Antp + CBs. Antp + CBs (yellow brackets) are neatly aligned in stereotypical 2x4 organization. (B’-F’) Schematics depicting relevant cell types in B-F: hematopoietic progenitors (magenta), PSC (green), and Antp + cardioblasts (blue). (G-J) Live-imaging stills from Hand-RFP,Antp-GAL4,10xUAS-myr:GFP embryos; RFP in magenta and GFP in white. (G) Timelapse shows dorsolateral view of PSC (magenta and white cells above green underline and below string of CBs, arrows) migrating as a collective from migration onset (0 m), when the PSC is lateral, to migration completion (around 2h50m), when the PSC is dorsal. Arrowhead indicates fixed positioning of the posterior-most PSC cell in the collective. (H-H’’’) Live-imaging stills show PSC protrusions during mid-migration. (H) Merge shows positional relation of PSC (white and magenta) and its protrusions to rest of lymph gland and CBs (along top of image). (H’-H’’’) Only GFP channel. Progressively shallower z-slices of same PSC. (H’) Anterior protrusions; some short (arrowheads) and one long (bracket) that encases lymph gland. (H’’) Posterior protrusion (bracket). (H’’’) Green dot indicates dorsal-most PSC cell with short, branching protrusions (arrowheads). Much of image contains Antp +epidermis. (I) Live-imaging stills of GFP channel only show PSC cells shifting position within collective. Initially connected yellow and cyan PSC cells (0 m, arrowhead) become separated by intercalation of magenta and green PSC cells while the posterior-most, dorsal vessel-adjacent PSC cell (orange dot) remains at a fixed position in the collective. (J) Same embryo as (G), GFP channel only. For the entire timelapse PSC cells migrate dorsally in close association with concurrently migrating neighbor tissues, CBs and Vm. Posterior-most PSC cell (arrowhead) maintains its dorsal vessel-adjacent position throughout. 0 min inset shows PSC proximity to Vm and 2hr 50min inset shows PSC proximity to CBs. (J’) Schematics depict relevant cell types for each panel of J time series: cardioblasts (blue), PSC (green), and visceral mesoderm (orange). (K) Live-imaging series from dorsal vantage point shows A1 AMs (magenta; labeled with org-1-HN39::RFP) contact PSCs (green outlines; labeled by Antp >CD8::GFP) throughout the timelapse as both tissues migrate to dorsal midline. (L) Schematics depict the relative positioning of the tissues identified to be near the PSC at migration onset (left) and completion (right).
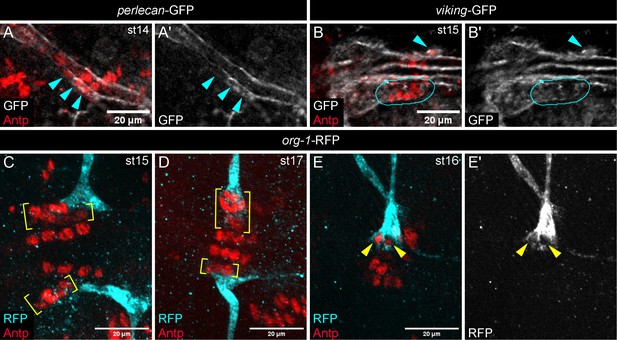
ECM components surround the PSC and AM encases PSC cells.
(A, B) ECM components Perlecan and Viking (Collagen IV subunit) surround migrating PSCs. (A) perlecan-GFP reporter, white, surrounds migrating Antp-labeled PSC cells, cyan arrowheads, of st14 embryo. (B) viking-GFP reporter, white, surrounds migrating PSC collective on embryo left side, cyan outline, and a single PSC cell on the right side, arrowhead. (A’, B’) GFP channel only. (C-E) org-1-RFP embryos with PSCs labeled by Antp, yellow brackets or arrowheads. (C) Alary muscle of first abdominal segment (A1 AM), cyan, contacts posterior of PSC collective in st15 embryo. (D) A1 AMs contact entire PSC collectives of st17 embryo. (E) st16 embryo with individual A1 AM fibers encasing two PSC cells, arrowheads. (E’) RFP channel alone. Scale bars as indicated.
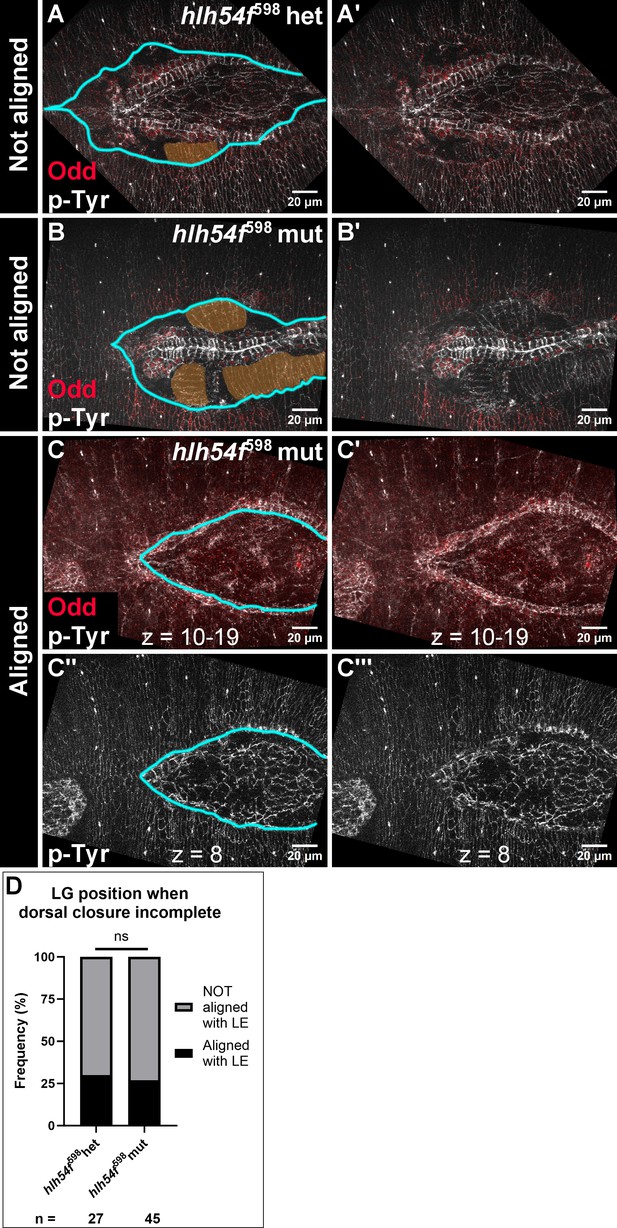
LG and CBs migrate independently of dorsal closure.
Analysis of LG alignment relative to leading edge epidermal cells in stage 17 embryos. Odd (red) marks LG and PSCs; p-Tyr (white) marks cell outlines. The specific cell contours, coupled to the depth of the slice imaged, can unequivocally identify and distinguish leading edge epidermal cells (LE), from CBs, and from visceral mesoderm, as well as other tissues. For this analysis, embryos were selected that exhibited a stalled LE, whether heterozygous (A) or homozygous (B, C) for hlh54f598. The LE front is marked by cyan-colored lines; and duplicate panels are shown without markup (A’, B’, C’, C’’’). Oftentimes, visceral mesoderm (false-colored orange) is visible when LGs and CBs have passed the LE (A, B). (A, A’) An example of a ‘Not aligned’ case in a heterozygote where the LG (Odd+, red nuclei) has migrated past the LE, and is visible more medially. (B, B’) An example of a ‘Not aligned’ case in a homozygous mutant. (C) An example of an ‘Aligned’ case in a homozygous mutant. (C, C’) A projection of nine Z-slices (4.5 µm) showing the LG and CBs. (C’’, C’’’) A single, shallower Z-slice focused on the stalled LE, aligned directly above the LG in (C, C’). (D) Quantification of LG alignment relative to stalled leading edge in hlh54f598 heterozygous and homozygous mutants. Ns, not significant; Fisher’s Exact Test. Scale bars as indicated. Sample sizes as indicated. There was no statistical difference between heterozygous and homozygous mutants in the frequency of LG-LE alignment upon incomplete dorsal closure, indicating that LG ability to migrate independently is unrelated to the hlh54f mutation itself.
Timelapse imaging of wildtype PSC migration from dorsolateral vantage visualized with Hand-RFP, Antp>myrGFP.
The PSC moves from its point of specification to its final position as a coalesced collective of cells. Stills from timelapse shown in Figure 1G.
Live-imaging of PSC collective visualized with Antp>myrGFP from dorsolateral vantage point during migration.
One PSC cell (cyan fill; orange dot) maintains a fixed positioning, adjacent to CBs, throughout the migration. The lateralmost PSC cells (magenta fill) shift position within the collective. Stills from timelapse shown in Figure 1I.
Timelapse imaging of wildtype PSC migration from dorsolateral vantage visualized with Hand-RFP,686 Antp>myrGFP (left) and Antp>myrGFP only (right).
The PSC (green), CBs (blue), and Vm (orange) migrate687 towards the dorsal midline in synchrony.
Magnified version of Figure 1—video 3.
Stills from timelapse shown in Figure 1J.
Live-imaging with dorsal view of PSC and AM migration visualized with Antp>mCD8:GFP and org-1-HN39::RFP, respectively, beginning at migration onset.
A1 AMs contact the PSC throughout the migration. Stills from timelapse shown in Figure 1K.
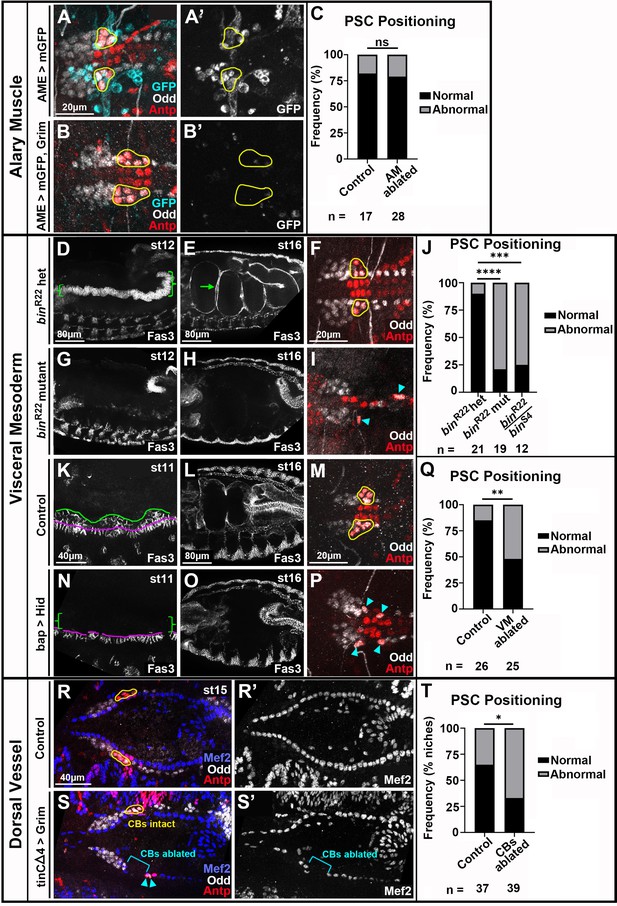
Visceral mesoderm and cardioblasts are required for PSC formation.
All PSCs, co-labeled by Antp and Odd, are viewed dorsally from st16 or 17 embryos unless otherwise noted. (A, B) LGs with normally-positioned PSCs outlined in yellow from control (A) and AM-ablated (B) embryos with AMs labeled by AME-Gal4 driven mCD8:GFP. (A’, B’) GFP channel only with PSC outlines overlayed. (C) PSC positioning quantification. (D, E) bin heterozygotes labeled with Fas3, lateral views. (D) Green bracket indicates st12 Vm. (E) st16 with green arrow indicating first Vm constriction that segregates two sections of gut. (F) bin heterozygote LGs with normal PSC positioning. (G, H) lateral views of bin mutants with minimal (G, st12) and absent (H, st16) Vm. (I) bin mutant LGs with dispersed PSCs (cyan arrowheads). (J) PSC positioning quantification including analysis of binR22/binS4 transheterozygous mutants. (K-P) control embryos (K-M) compared to Vm-ablated embryos (N-P). (K) Lateral view of normal st11 Vm labeled by Fas3. Founder cells are below magenta line and fusion competent myoblasts are between magenta and green lines. (L) Dorsal view of normal st16 Vm. (M) Normal PSC positioning, yellow outlines, in control. (N) Fusion competent myoblasts absent, green brackets, in st11 Vm ablated embryo. Most founder cells present with occasional gaps. (O) Vm absent in st16 Vm ablated embryo. (P) Abnormal PSC positioning, cyan arrowheads. (Q) PSC positioning quantification. (R, S) st15 LGs with cardioblasts labeled by Mef2 in control (R) and CB-ablated embryos (S). (R) Normal PSC positioning, yellow outlines. (S) CBs ablated on left side, bracket, and corresponding PSC is mis-positioned, arrowheads, while right side CBs are intact and the R PSC is positioned normally, yellow outline. (R’, S’) Mef2 channel only. (T) Quantification comparing PSC positioning in control embryos to positioning of PSCs with ablated ipsilateral CBs. Scale bars as indicated. Ns = not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, Fisher’s Exact test. Sample sizes as indicated.
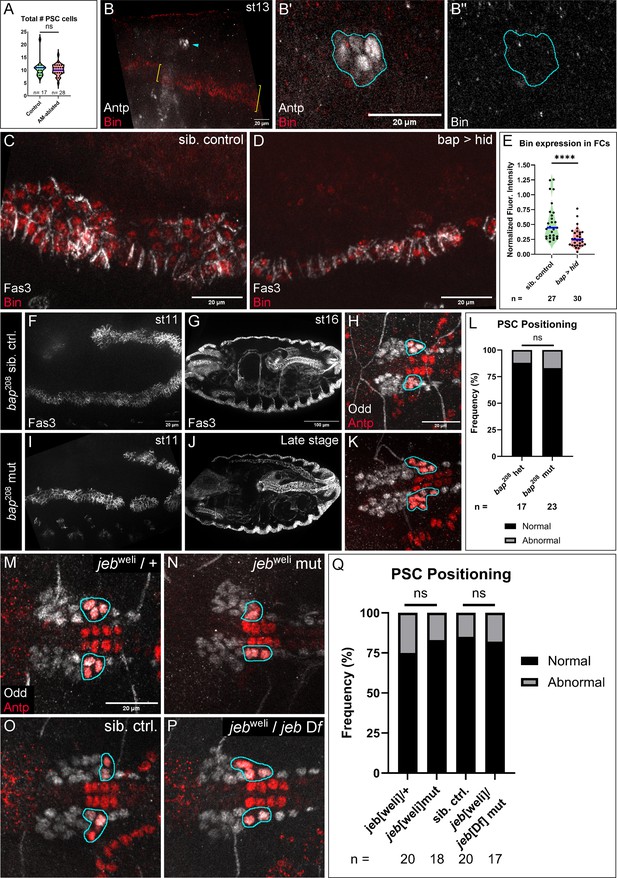
PSC analysis under various manipulations.
All PSC positioning analyses were completed on st16 or st17 embryos by co-labeling with Antp and Odd. (A) Quantitation of PSC cells in controls and AM-ablated embryos. Not significant (ns), Mann-Whitney test. (B) Lateral view of st13 embryo stained for Antp (white) and Bin (red). Antp labels PSC collective (arrowhead) and Bin is expressed in Vm (brackets), as expected. (B’) Magnified view of Antp+ collective (outline) shows no detectable Bin. (B’’) Bin channel only; PSC collective outlined. (C, D) Fas3-labeled Vm (white) of st11 control (C) and FCM-ablated (D) embryos also stained for Bin (red). (E) Bin expression is reduced in remaining FCs of Vm-ablated embryos. ****p<0.0001, Mann-Whiteny test. (F-K) Analysis of bap208 sibling controls (F-H) and hypomorphic mutants (I-K). (F, I) Lateral view of st11 Fas3-labeled Vm precursors of control (F) and mutant (I), which has almost normal Vm organization with occasional gaps. (G) Dorsolateral view of Fas3 stained st16 sibling control embryo with normal Vm morphology. (H) Sibling control LGs with normal PSCs. (J) Dorsolateral view of Fas3 stained late stage mutant embryo with disorganized Vm that is missing in the midline region where the PSC is normally located. (K) bap208 mutant LGs with normal PSCs. (L) Quantification of frequency of PSC positioning phenotype in bap208 sibling controls and mutants; ns, Fisher’s Exact test. (M-Q) analysis of PSC positioning for various jeb alleles; frequency of phenotypes quantified in Q (ns, Fisher’s Exact test). (M, N) LGs with normal PSC positioning from jebweli heterozygous (M) and homozygous (N) mutant. (O, P) LG with normal PSC positioning from a sibling control (O) and a jebweli/jeb Df transheterozygous mutant (P). Scale bars as indicated. Sample sizes as indicated.
-
Figure 2—figure supplement 1—source data 1
PSC cell count in Control and AM-ablated cases.
- https://cdn.elifesciences.org/articles/100455/elife-100455-fig2-figsupp2-data1-v1.xlsx
-
Figure 2—figure supplement 1—source data 2
Bin fluorescence intensity measurements in Control and Ablated.
- https://cdn.elifesciences.org/articles/100455/elife-100455-fig2-figsupp2-data2-v1.xlsx
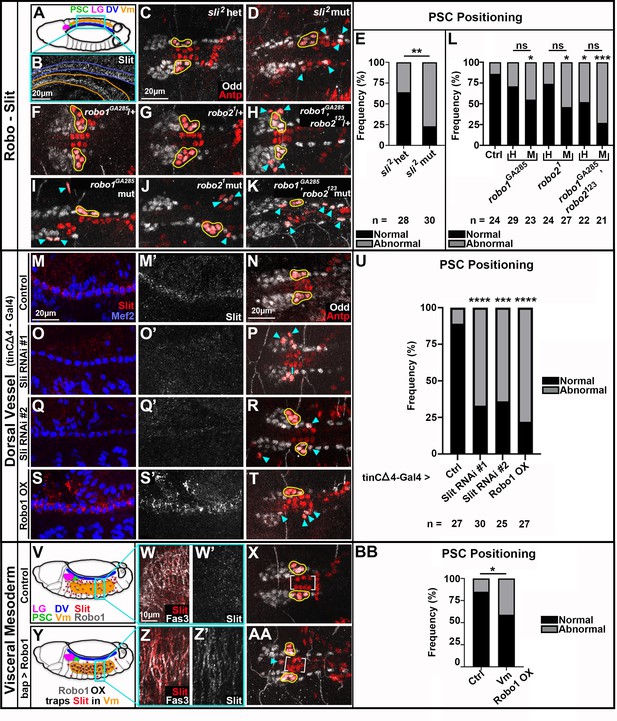
Slit from Vm and CBs signals through Robo for PSC positioning.
All PSCs, co-labeled by Antp and Odd, are viewed dorsally from st16 or 17 embryos. Normally coalesced and positioned PSC cells are outlined in yellow; dispersed PSC cells indicated by cyan arrowheads (A) Schematic of dorsal st14 embryo. Relevant tissues are indicated; Vm is ventral to the rest. (B) Slit expression in st14 CBs, blue outline, and Vm, orange outline; these tissues are in reversed orientation from the schematic due to embryo tilt, and the projection necessary to have both tissues in view. (C-E) Analysis of PSC positioning in sli2 heterozygotes (C, normal PSCs) and sli2 mutants (D, dispersed PSCs), with frequency of positioning phenotype quantified in (E. F-L) Analysis of PSC positioning under various robo depletion scenarios. robo1 heterozygote (F) and robo2 heterozygote (G) with normal PSCs. (H) robo1,robo2 double heterozygote with abnormal PSCs. robo1 single (I), robo2 single (J), and robo1, robo2 double (K) mutants with abnormal PSCs; frequency of positioning phenotype quantified in L (‘H’) is heterozygote, (‘M’ is mutant). (M-U) Analysis of PSC positioning when Slit signaling from CBs is compromised; frequency of PSC positioning phenotype quantified in (U. M, O, Q, S) Slit expression in Mef2 labeled cardioblasts of controls (M) and tinCΔ4-GAL4 driven Slit RNAi knockdown (O, Q) or Robo1 overexpression (S) embryos. (M’, O’, Q’, S’) Slit channel only. (N) Control with normal PSCs. (P, R, T) Dispersed PSCs upon compromised Slit signaling from CBs. V-BB Analysis of PSC positioning when Slit signaling from Vm is compromised; frequency of PSC positioning phenotype quantified in BB. (V, Y) Schematics of dorsolateral st14 control (V) or bap-GAL4 driven Robo1 overexpression (Y) embryos with relevant tissues and proteins indicated. (W) Diffuse Slit expression in Fas3-labeled Vm of st14 control embryo. (Z) Slit trapping at Fas3-labeled Vm membranes in bap-GAL4-driven Robo1 overexpression embryo. (W’, Z’) Slit channel only. (X) Normally positioned PSCs and Antp+ CBs (brackets) in control. (AA) Abnormally positioned PSC and Antp+ CBs (brackets) in bap-GAL4-driven Robo1 overexpression embryo. Scale bars as indicated. Ns = not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, Fisher’s Exact test. Sample sizes as indicated.
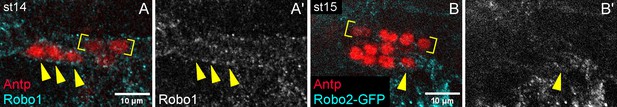
Robo1 and Robo2 are expressed by PSC cells.
(A) Robo1 expression (cyan) on PSC cell membranes (arrowheads) of st14 embryo stained for Antp; brackets are two of the eventual 4 Antp +CBs. (A’) Robo1 channel alone. (B) robo2-GFP reporter expression (cyan) on membrane of single PSC cell (arrowhead) of st15 embryo stained for Antp; brackets enclose Antp +CBs. (B’) GFP channel alone.
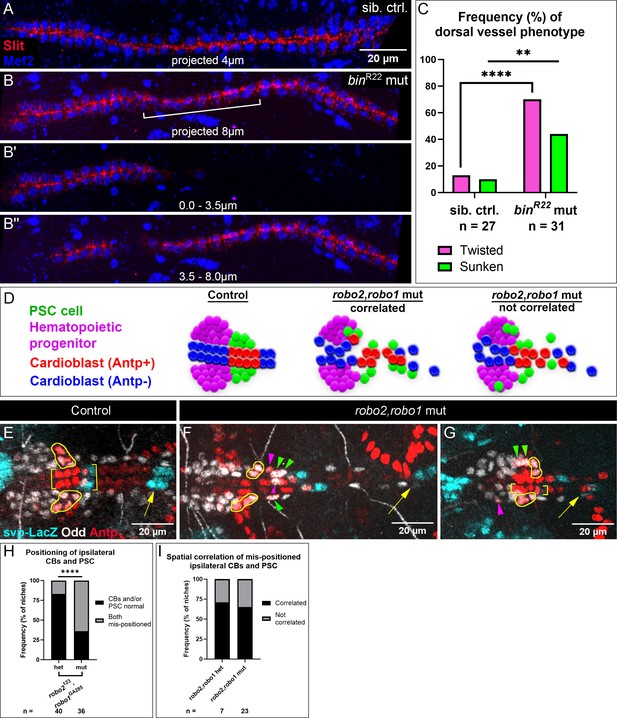
PSC positioning requires Vm-mediated dorsal vessel organization and Robo activation in PSC cells.
(A-C) Analysis of dorsal vessels with and without Vm; phenotype frequencies quantified in (C). (A, B) Mef2-labeled CBs and Slit-labeled lumens of dorsal vessels. (A) Dorsal vessel of sibling control, captured in 4.0 µm projection, shows normal, neatly-aligned CB organization. (B) binR22 mutant with twisted dorsal vessel (bracket), fully captured only by projection of an 8.0 µm deep stack. (B’) twisted phenotype evidenced by the absence of the sunken portion in the first 3.5 µm;. (B’’) 3.5–8 µm projection. (D) Schematics depicting three expectations for PSC and CB positioning. In controls (left), PSCs are coalesced and adjacent to CBs. In robo2,robo1 mutants, either PSC cells are associated with mis-positioned CBs (middle), suggesting passive displacement of the PSCs; or displaced PSC cells are sometimes separate from mis-positioned CBs (right), suggesting the PSC cells themselves require activated Robo signaling. (E-I) Analysis of CB and PSC positioning with additional CB marker, svp-LacZ. Brackets indicate normally-positioned CBs. Yellow outlines indicate normally-positioned PSC cells. Arrowheads indicate abnormally-positioned PSC cells; green arrowheads indicate passive mis-positioning and magenta arrowheads are displaced PSC cells without a similarly displaced CB nearby. Arrows indicate the next set of svp +CBs to the posterior. (E) Control with normally-positioned PSCs and CBs; second set of svp +CBs (arrow) nearby. (F) robo2,robo1 mutant with abnormal CB positioning; Antp +CBs are dispersed and the second set of svp +CBs (arrow) are displaced far posteriorly. PSCs display both passive mis-positioning posteriorly (green arrowheads) and mis-positioning without a similarly mispositioned CB nearby magenta arrowheads; bottom PSC cell invaded midline and top PSC cell is displaced laterally. (G) robo2,robo1 mutant with left side CBs positioned normally (brackets) and the second set of svp +CBs in view (arrow); despite normal CB positioning, the ipsilateral PSC has a mis-positioned cell (magenta arrowhead). Right side CBs positioned abnormally (an Antp +CB is displaced laterally and the second set of svp +CBs are displaced posteriorly, not in view); correspondingly, the ipsilateral PSC has two laterally displaced cells (green arrowheads). (H) Frequency of mis-positioning of both a PSC and the ipsilateral CBs; this occurs more frequently in robo2,robo1 mutants than heterozygotes. (I) Frequency of correlated mis-positioning of PSCs and CBs. Both robo2,robo1 heterozygotes and mutants have CB-independent instances of PSC mis-positioning. Sample sizes as indicated. **p<0.01, ****p<0.0001, Fisher’s Exact test. Scale bars as indicated.
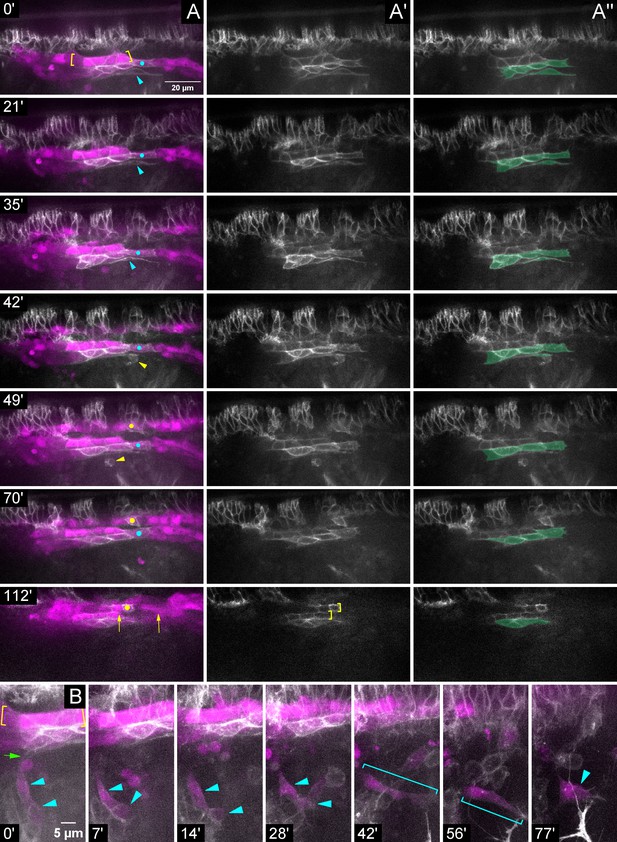
During migration Slit is required for proper PSC adhesions to CBs and for association of the PSC collective.
Stills from timelapse imaging of sli mutant embryo with LG and PSC labeled by Hand-RFP, Antp >myrGFP; dorsolateral view. (A) Multichannel, (A’) single Antp >myrGFP channel, or (A’’) single Antp >myrGFP channel false-colored for PSC cells; time increasing vertically (see timestamps). (A) The line of GFP+ cells within the brackets are Antp+ CBs; GFP+ cells below this are Antp+ PSC cells (see A’’). Elongated PSC cell (cyan arrowhead; 0–35’) cleared by macrophage (yellow arrowhead; 42–49’). Anterior and posterior edges of another PSC cell (cyan dot) bridges separated CBs (0–70’); same PSC cell aberrantly contacts a contralateral Antp+ CB (yellow dot; 49–70’). Persisting gap in CBs (arrows; 112’) evident in same region. (A’) Single Antp >myrGFP channel. Brackets indicate mis-aligned contralateral Antp+ CBs (112‘). (B) Stills with time increasing across the row, revealing a different aspect of the same sli mutant embryo in (A). Cells within the yellow brackets are Antp+ CBs. The PSC contains two laterally displaced PSC cells (cyan arrowheads; 0–28’) barely attached (green arrow; 0’) to main cluster. These cells are disconnected from the main cluster (7’) and remain stationary as the other PSC cells and CBs migrate away (7–77’). The disconnected PSC cell(s) change shape (cyan brackets) and develop membranous spikes (77’ arrowhead). Scale bars as indicated.
Live-imaging with dorsolateral view of PSC migration in sli2 mutant visualized with Hand-RFP, Antp>myrGFP (left) or Antp>myrGFP only (right) beginning midway in the migration.
One PSC cell elongates and is phagocytosed; another makes aberrant contact with contralateral CBs. Stills from timelapse shown in Figure 5A.
Live-imaging with dorsolateral view of PSC migration in sli2 mutant visualized with Hand-RFP, Antp>myrGFP.
Two PSC cells become separated from the main cluster as it migrates away dorsally. Stills from timelapse shown in Figure 5B.
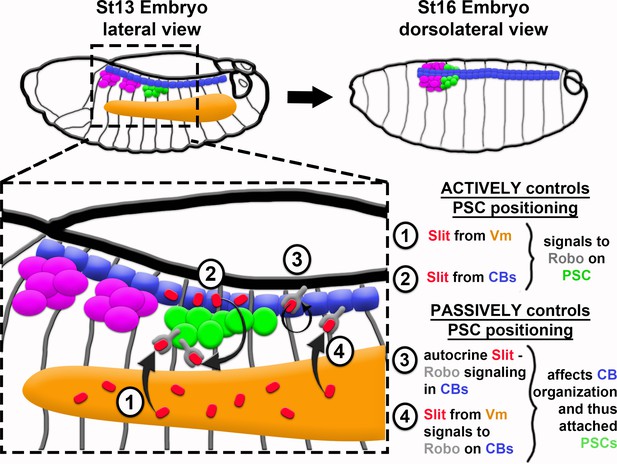
Model of PSC formation.
PSC migration from its point of specification, laterally in the embryo (left), to its final position at the dorsal midline (right), requires both active Robo signaling in PSC cells (1 and 2) and proper organization of CBs – a passive, indirect control of PSC positioning (3 and 4). Slit from VM (1) and from CBs (2) directly impacts PSC positioning by binding to Robo1 and Robo2 receptors on PSC cells. In a more passive manner, Slit controls PSC positioning via binding to Robo receptors on CBs, which ensures their proper polarity and organization both by autocrine signaling in CBs (3; previously known) and by way of Slit emanating from Vm (4; novel finding from this work).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | Antp-GAL4 | Emerald and Cohen, 2004 | FLYB:FBal0155891 | FlyBase symbol: GAL4Antp-21 |
| Genetic reagent (D. melanogaster) | w1118 | Bloomington Drosophila Stock Center | BDSC:3605; FLYB:FBal0018186; RRID:BDSC_3605 | FlyBase symbol: w1118 |
| Genetic reagent (D. melanogaster) | Hand-RFP | other | Gift from Georg Vogler | |
| Genetic reagent (D. melanogaster) | UAS-myr:GFP | Bloomington Drosophila Stock Center | BDSC:32200; FLYB:FBti0131976 RRID:BDSC_32200 | FlyBase symbol: P{10XUAS-IVS-myr::GFP}su(Hw)attP1 |
| Genetic reagent (D. melanogaster) | tupAME-GAL4 | Bataillé et al., 2020 | FLYB: FBtp0142468 | FlyBase symbol: P{tup-GAL4.AME-R} Gift from J.L. Frendo |
| Genetic reagent (D. melanogaster) | UAS-CD8:GFP | other | Gift from J.L. Frendo | |
| Genetic reagent (D. melanogaster) | org-1-HN39-RFP | Schaub et al., 2015 | FLYB:FBal0276776 | FlyBase symbol: RFPorg-1.HN39 |
| Genetic reagent (D. melanogaster) | UAS-grim | Hugo Bellen | FLYB:FBti0154788 | Flybase symbol: Dmel\P{UAS-grim.Y}2 |
| Genetic reagent (D. melanogaster) | binR22 | Zaffran et al., 2001 | FLYB:FBal0043738 | Flybase symbol: Dmel\binR22 |
| Genetic reagent (D. melanogaster) | binS4 | Zaffran et al., 2001 | FLYB:FBal0043739 | Flybase symbol: Dmel\binS4 |
| Genetic reagent (D. melanogaster) | UAS-hid | Bloomington Drosophila Stock Center | BDSC:65403; FLYB:FBti0183136 RRID:BDSC_65403 | Flybase symbol: Dmel\P{UAS-hid.Z}2 |
| Genetic reagent (D. melanogaster) | bap-GAL4 | Zaffran et al., 2001 | BDSC:91540; FLYB:FBti0214156 | Flybase symbol: Dmel\P{bap-GAL4.3}1.1 |
| Genetic reagent (D. melanogaster) | bap-GAL4 | other | gift from Manfred Frasch; Chr: X | |
| Genetic reagent (D. melanogaster) | tinCΔ4-GAL4 | Bloomington Drosophila Stock Center | BDSC:92965; FLYB:FBti0216630 | Flybase symbol: Dmel\P{tinC-Gal4.Δ4}12 a |
| Genetic reagent (D. melanogaster) | slit2 | Bloomington Drosophila Stock Center | BDSC:3266 FLYB:FBal0015700 | Flybase symbol: Dmel\sli2 |
| Genetic reagent (D. melanogaster) | robo1GA285 | other | FLYB:FBal0032588 | Gift from Greg Bashaw Flybase symbol: Dmel\robo11 |
| Genetic reagent (D. melanogaster) | robo21 | Rajagopalan et al., 2000 | FLYB:FBal0121562 | Gift from Greg Bashaw Flybase symbol: Dmel\robo21 |
| Genetic reagent (D. melanogaster) | robo2123 | other | FLYB:FBal0123720 | Gift from Greg Bashaw Flybase symbol: Dmel\robo2X123 |
| Genetic reagent (D. melanogaster) | UAS-Slit RNAi #1 | Vienna Drosophila Stock Center | VDRC:v108853 FLYB:FBti0159991 | Flybase symbol: Dmel\P{KK100803}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-Slit RNAi #2 | Bloomington Drosophila Stock Center | BDSC:31468 FLYB:FBal0245521 | Flybase symbol: Dmel\sliJF01229 |
| Genetic reagent (D. melanogaster) | UAS-Robo1 OX | Evans et al., 2015 | BDSC:97240 FLYB:FBal0316479 | Flybase symbol: Dmel\robo1 ΔC.10xUAS.Tag:HA,Tag:SS(wg) |
| Genetic reagent (D. melanogaster) | UAS-dcr2 | Bloomington Drosophila Stock Center | BDSC:24650 FLYB:FBti0100275 | Flybase symbol: Dmel\P{UAS-Dcr-2.D}2 |
| Genetic reagent (D. melanogaster) | UAS-dcr2 | Bloomington Drosophila Stock Center | BDSC:24651 FLYB:FBti0100276 | Flybase symbol: Dmel\P{UAS-Dcr-2.D}10 |
| Genetic reagent (D. melanogaster) | svp-lacZ | Bloomington Drosophila Stock Center | BDSC:7314 FLYB:FBti0002862 | Flybase symbol: Dmel\P{HZ}svp3 |
| Genetic reagent (D. melanogaster) | perlecan-GFP | Flytrap; GFP Protein Trap Database | FLYB:FBal0243609 | Flybase symbol: Dmel\trolZCL1700 |
| Genetic reagent (D. melanogaster) | viking-GFP | Buszczak et al., 2007 | FLYB:FBal0211825 | Flybase symbol: Dmel\vkgCC00791 |
| Genetic reagent (D. melanogaster) | bap208 | Bloomington Drosophila Stock Center | BDSC:91539 FLYB:FBal0034201 | Flybase symbol: Dmel\bap208 |
| Genetic reagent (D. melanogaster) | jebweli | Stute et al., 2004 | FLYB:FBal0159133 | Flybase symbol: Dmel\jebweli |
| Genetic reagent (D. melanogaster) | jeb Df | Bloomington Drosophila Stock Center | BDSC:26551 FLYB:FBab0045764 | Flybase symbol: Df(2 R)BSC699 |
| Genetic reagent (D. melanogaster) | robo2-GFP | Bloomington Drosophila Stock Center | BDSC:61774 FLYB:FBal0265307 | Flybase symbol: Dmel\robo2MI04295 |
| Antibody | anti-Antp (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat#:8C11, RRID:AB_528083 | IF(1:50) |
| Antibody | anti-Odd skipped (Rabbit polyclonal) | Ward and Skeath, 2000 | IF(1:400); gift from James Skeath | |
| Antibody | anti-GFP (Chick polyclonal) | Aves labs | Cat#:GFP-1020 RRID:AB_2307313 | IF(1:1500) |
| Antibody | anti-Fas3 (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat#:7G10 RRID:AB_528238 | IF(1:50) |
| Antibody | anti-Mef2 (Rabbit polyclonal) | Developmental Studies Hybridoma Bank | Cat#:Mef2 RRID:AB_2892602 | IF(1:1000) |
| Antibody | anti-Slit (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat#:C555.6D RRID:AB_528470 | IF(1:200); gift from Greg Bashaw |
| Antibody | anti-LacZ (Chick polyclonal) | Abcam | Cat#:ab9361 RRID:AB_307210 | IF(1:1000) |
| Antibody | anti-RFP (Rabbit polyclonal) | Abcam | Cat#:ab62341 RRID:AB_945213 | IF(1:1000) |
| Antibody | anti-Bin (Rabbit polyclonal) | other | IF(1:100); gift from Eileen Furlong | |
| Antibody | anti-Robo1 (Mouse monoclonal) | other | IF(1:200); gift from Greg Bashaw | |
| Antibody | anti-Odd skipped (Guinea pig polyclonal) | other | IF(1:1200); gift from John Reinitz | |
| Chemical compound, drug | Paraformaldehyde | Electron Microscopy Sciences | Cat#:15710 | |
| Chemical compound, drug | Propyl-gallate | Sigma Aldrich | PubChem Substance ID:24898394; SKU:P3130; CAS Number:121-79-9 | |
| Chemical compound, drug | Normal Donkey Serum | Jackson ImmunoResearch Labs Inc | Cat#:017-000-121 RRID:AB_2337258 | |
| Chemical compound, drug | Ringer’s solution | other | Recipe from de Cuevas and Spradling, 1998 | |
| Chemical compound, drug | Triton X-100 | MilliporeSigma | CAS Number: 9036-19-5 | |
| Software, algorithm | FIJI | ImageJ | RRID:SCR_002285 | http://fiji.sc |
| Software, algorithm | Photoshop | Adobe | RRID:SCR_014199 | https://www.adobe.com/products/photoshop.html |
| Software, algorithm | Prism | Graphpad | RRID:SCR_002798 | v9.0.0-v10.0.0 |
| Software, algorithm | Axio-Vision Imaging Software | Zeiss | v4.8.1 | |
| Software, algorithm | VisiView | Visitron | ||
| Software, algorithm | Metamorph Microscopy Automation and Image Analysis Software | Leica | v7.8.40 | |
| Other | 63 x / 1.2 NA water immersion objective | Leica | ||
| Other | 60 x / 1.3 NA silicone immersion objective | Olympus | ||
| Other | AxioCam HRm | Zeiss | ||
| Other | 40 x / 1.2 NA water immersion objective | Zeiss | ||
| Other | 20 x / 0.8 NA objective | Zeiss | ||
| Other | M165FC | Leica | ||
| Other | Achromat 1.6 x objective | Leica | ||
| Other | GFP Filter set ET470/40 x; ET525/50 m | Leica | ||
| Other | mCherry Filter set ET560/40 x; ET630/75 m | Leica | ||
| Other | pco.edge 4.2 bi sCMOS | PCO | ||
| Other | Cell Center Stockroom (Penn) | other | RRID:SCR_022399 | |
| Other | CDB Microscopy Core (Penn) | other | RRID:SCR_022373 |





