Phase transition of WTAP regulates m6A modification of interferon-stimulated genes
Figures
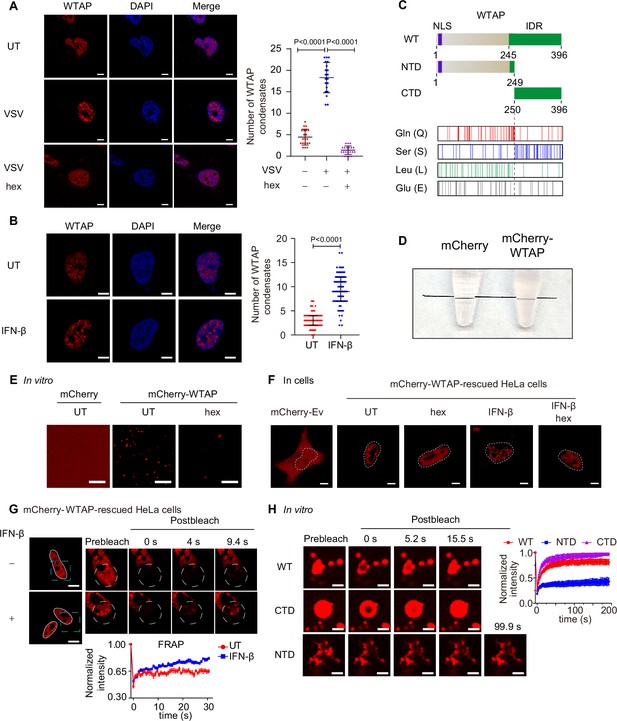
Wilm’s tumor-associated protein (WTAP) goes through phase transition with IFN-β stimulation.
(A) THP-1-derived macrophages were infected with Vesicular Stomatitis Virus (VSV) (m.o.i.=0.1) for 24 hr together with or without 5% 1,6-hexanediol (hex) and 10 μg/mL digitonin for 2 hr or left untreated (UT). Endogenous WTAP was stained and imaged using confocal microscopy. The number of WTAP condensates that diameter over 0.4 μm of n=20 cells were counted through ImageJ and shown. Scale bars indicated 5 μm. (B) HeLa cells were placed on the dishes and stimulated with or without 10 ng/mL IFN-β for 1 hr at 37°C. Endogenous WTAP was stained and imaged using confocal microscopy. The number of WTAP condensates that diameter over 0.4 μm of n=80 cells were counted through ImageJ and shown. Scale bars indicated 5 μm. (C) Domain structures (top) and distribution of amino acids (bottom) of WTAP protein. WT, wild-type. NTD, N-terminal domain. CTD, C-terminal domain. NLS, nuclear localization signal. IDR, intrinsically disordered regions. Gln, Glutamine. Ser, Serine. Lue, Leucine. Glu, Glutamic acid. (D) Tubes containing physiological buffer with recombinant mCherry (10 μM) or mCherry-WTAP (10 μM) were compared, in which recombinant mCherry-WTAP underwent phase separation. (E) Foci formation of recombinant mCherry-WTAP (10 μM) with or without 5% 1,6-hexanediol (hex) in vitro was observed through confocal microscopy. Scale bars indicated 5 μm. (F) Phase separation of mCherry-WTAP in mCherry-WTAP-rescued HeLa cells treated with or without 5% hex and 20 μg/mL digitonin were observed through confocal microscopy. Representative images of n=20 cells were shown. Scale bars indicated 5 μm. (G) mCherry-WTAP-rescued HeLa cells were placed on dishes and treated with or without 10 ng/mL IFN-β for 1 hr at 37℃. After stimulation, bleaching of the WTAP foci was performed and quantification of fluorescence recovery after photobleaching (FRAP) of mCherry-WTAP aggregates was analyzed. The start time point of recovery after photobleaching was defined as 0 s. Representative images of n=10 cells were shown. Scale bars indicated 5 μm. (H) Recombinant mCherry-WT WTAP, N-terminal domain (NTD), and C-terminal domain (CTD) (10 μM) were mixed with the physiological buffer and placed on the dishes at 37℃ (Prebleach). After incubation, bleaching was performed and quantification of FRAP of recombinant mCherry-WT WTAP, NTD, and CTD were analyzed. Representative images of n=6 condensates were shown, and the normalized intensity was measured and analyzed. The start time point of recovery after photobleaching was defined as 0 s. Scale bars indicated 2 μm. All error bars, mean values ± SD, p-values were determined by unpaired two-tailed Student’s t-test of n=20 cells in (A) and n=80 cells in (B). For (A, B, D–H), similar results were obtained for three independent biological experiments.
-
Figure 1—source data 1
Numerical data used to generate Figure 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig1-data1-v1.xlsx
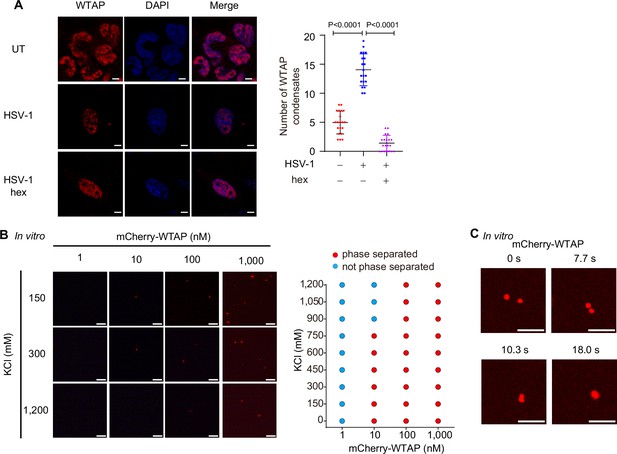
Wilm’s tumor-associated protein (WTAP) undergoes phase separation.
(A) THP-1-derived macrophages were infected with Herpes Simplex Virus-1 (HSV-1) (m.o.i.=1) for 24 hr together with or without 5% 1,6-hexanediol (hex) and 10 μg/mL digitonin for 2 hr or left untreated (UT). WTAP were stained and imaged using a confocal microscope. The number of WTAP condensates that diameter over 0.4 μm of n=20 cells were counted through ImageJ and shown. Scale bars indicated 5 μm. (B) Phase separation of recombinant mCherry-WTAP in different concentrations incubated with different concentrations of KCl were observed through confocal microscopy. Representative images were shown (left), and phase separation diagram was listed (right). Scale bars indicated 10 μm. (C) Recombinant mCherry-WTAP (10 μM) was mixed with physiological buffer and incubated at 37℃. Time-lapse microscopy of merging condensates was performed and representative images were shown. The beginning of our observation by confocal microscopy was defined as 0 s. Scale bars indicated 5 μm. All error bars, mean values ± SD, p-values were determined by unpaired two-tailed Student’s t-test of n=20 cells in (A). For (B, C), similar results were obtained for three independent biological experiments.
-
Figure 1—figure supplement 1—source data 1
Numerical data used to generate Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig1-figsupp1-data1-v1.xlsx
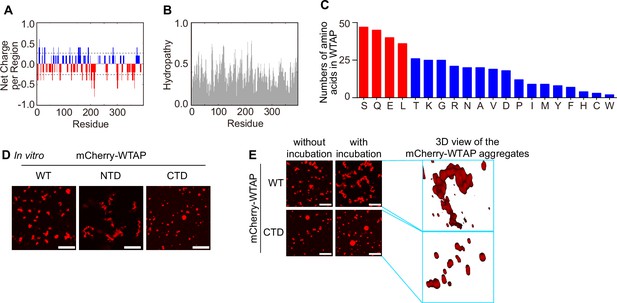
Serine-rich C-terminal domain (CTD) and glutamine-rich N-terminal domain (NTD) of Wilm’s tumor-associated protein (WTAP) provide the potential of aggregates or liquid droplets of WTAP, respectively.
(A, B) Net charge per residue (A) and hydropathy (B) analysis of WTAP was performed through CIDER predictor (http://pappulab.wustl.edu/CIDER/analysis/). (C) Numbers of different amino acids within WTAP was calculated and shown. Four abundant amino acids (Serine (S), glutamine (G), glutamic acid (E) and leucine (L)) were marked as red while the rest were labeled as blue. (D) Phase separation of recombinant mCherry-WTAP WT, N-terminal domain (NTD), and CTD (40 μM) were observed through confocal microscopy. Scale bars indicated 10 μm. (E) Recombinant mCherry-WTAP (10 μM) was mixed with physiological buffer and incubated at 37℃. Time-lapse microscopy of merging condensates were performed and representative images were shown. The beginning of our observation by confocal microscopy was defined as 0 s. Scale bars indicated 10 μm. For (D, E), similar results were obtained for three independent biological experiments.
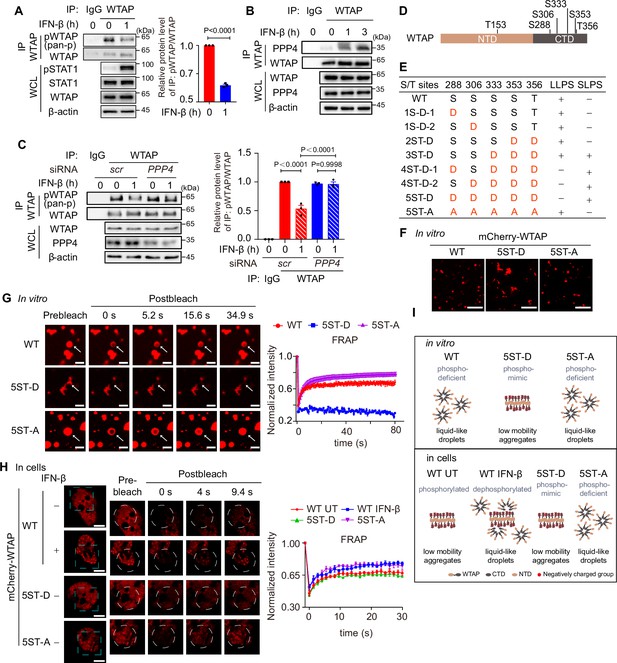
IFN-β-mediated dephosphorylation of Wilm’s tumor-associated protein (WTAP) induces its phase transition.
(A) THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 1 hr or left untreated. Whole cell lysate (WCL) was collected and immunoprecipitation (IP) experiment using anti-WTAP antibody or rabbit IgG was performed, followed by immunoblot. pWTAP was detected by anti-phosphoserine/threonine/tyrosine antibody (pan-p). Relative protein level was shown. (B) THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 1 hr or left untreated. WCL was collected and IP experiment using anti-WTAP antibody or rabbit IgG was performed, followed by immunoblot. (C) THP-1-derived macrophages were transfected with scrambled (scr) or PPP4-targeted siRNA and stimulated with or without 10 ng/mL IFN-β for 1 hr. WCL was collected and IP experiment with anti-WTAP antibody or rabbit IgG was performed, followed by immunoblot. pWTAP was detected by anti-phosphoserine/threonine/tyrosine antibody (pan-p). Relative protein level was measured and analyzed. (D) mCherry-WTAP was purified from HEK 293T cells expressing mCherry-WTAP using anti-WTAP antibody and analyzed by mass spectrometry (MS) for the phosphorylation. Six phosphorylated residues were identified. Data of MS assay for five phosphorylated sites within CTD were shown in Figure 2—figure supplement 1D. (E, F) Recombinant mCherry-WTAP mutants with indicated serine/threonine (S/T) to aspartate (D) or alanine (A) mutation were listed in (E), while representative images of the phase-separated mCherry-WT WTAP, 5ST-D and 5ST-A mutants were shown in (F). Images of phase separation of other mCherry-WTAP mutants (10 μM) were shown in Figure 2—figure supplement 1E. Scale bars indicated 5 μm. (G) Recombinant mCherry-WT WTAP (10 μM), 5ST-D mutant (10 μM) or 5ST-A mutant (10 μM) was mixed with the physiological buffer and placed on the dishes at 37℃ (Prebleach). After incubation, bleaching was performed and quantification of fluorescence recovery after photobleaching (FRAP) of recombinant mCherry-WT WTAP, 5ST-D, or 5ST-A mutant were analyzed. Representative images of n=5 condensates were shown and the normalized intensity was measured and analyzed. The start time point of recovery after photobleaching was defined as 0 s. Arrow indicated the FRAP area while scale bars indicated 2 μm. (H) mCherry-WT WTAP, 5ST-D, or 5ST-A mutant-rescued HeLa cells were placed on the dishes and stimulated with or without 10 ng/mL IFN-β for 1 hr at 37℃. After seeding, bleaching of the WTAP foci was performed and quantification of FRAP of mCherry-WTAP aggregates was analyzed. Representative images of n=5 cells were shown, and the normalized intensity was measured and analyzed. The start time point of recovery after photobleaching was defined as 0 s. Scale bars indicated 5 μm. (I) Schematic figure of phase separation of WT WTAP, 5ST-D, and 5ST-A mutants in vitro (above) and WT WTAP with IFN-β stimulation or left untreated (UT), 5ST-D and 5ST-A mutants in cells (below). All error bars, mean values ± SD, p-values were determined by unpaired two-tailed Student’s t-test of n=3 independent biological experiments in (A, C). For (A–C, E–H), similar results were obtained for three independent biological experiments.
-
Figure 2—source data 1
PDF files containing original figures of immunoblot analysis displayed in Figure 2, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig2-data1-v1.zip
-
Figure 2—source data 2
Original files for immunoblot analysis displayed in Figure 2.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig2-data2-v1.zip
-
Figure 2—source data 3
Numerical data used to generate Figure 2.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig2-data3-v1.xlsx
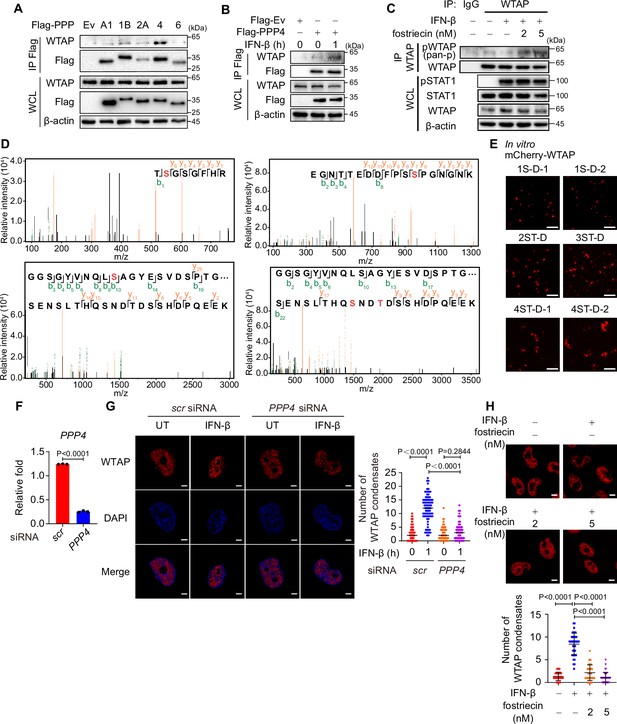
Protein phosphatases 4 (PPP4) is responsible for IFN-β-induced dephosphorylation of Wilm’s tumor-associated protein (WTAP).
(A) HEK 293T cells transfected with Flag-tagged empty vector (Ev) or protein phosphatases (PPPs). Whole cell lysate (WCL) was collected and immunoprecipitation (IP) experiment with anti-Flag beads was performed, followed by immunoblot. (B) HEK 293T cells transfected with Flag-tagged Ev or PPP4, followed by IFN-β or left untreated. WCL was collected and IP experiment with anti-Flag antibody was performed, followed by immunoblot. (C) THP-1-derived macrophages were pre-treated with 2 nM or 5 nM fostriecin for 24 hr or left untreated, followed by 10 ng/mL IFN-β for 1 hr or left untreated. WCL was collected and IP experiment using anti-WTAP antibody or rabbit IgG was performed, followed by immunoblot. pWTAP was detected by anti-phosphoserine/threonine/tyrosine antibody (pan-p). (D) Data of mass spectrometry (MS) assay of the five phosphorylated sites within WTAP CTD. (E) Representative images of the phase-separated mCherry-WTAP mutants (containing different numbers of serine (S)/threonine (T) to aspartic acid (D) mutants). Scale bars indicated 5 μm. (F) THP-1-derived macrophages were transfected with scrambled (scr) or PPP4-targeted siRNA. Expression of PPP4 was detected by quantitative real-time polymerase chain reaction (qPCR) assay after siRNA transfection for 48 hr. (G) THP-1-derived macrophages were transfected with scrambled (scr) or PPP4-targeted siRNA and stimulated with or without 10 ng/mL IFN-β for 1 hr. Endogenous WTAP was stained and imaged using confocal microscopy. The number of WTAP condensates that diameter over 0.4 μm of n=80 cells were counted through ImageJ and shown. Scale bars indicated 5 μm. (H) mCherry-WTAP-rescued HeLa cells were pre-treated with 2 nM or 5 nM fostriecin for 24 hr or left untreated, followed by 10 ng/mL IFN-β for 1 hr or left untreated. Phase separation of mCherry-WTAP was observed through confocal microscopy. The number of WTAP condensates that diameter over 0.4 μm of n=40 cells were counted through ImageJ and shown. Scale bars indicated 5 μm. All error bars, mean values ± SD, p-values were determined by two-way ANOVA test of n=80 cells in (G) and unpaired two-tailed Student’s t-test of n=40 cells in (H). All error bars, mean values ± SEM, p-values were determined by unpaired two-tailed Student’s t-test of n=3 independent biological experiments in (F). For (A–C, E, G, H), similar results were obtained for three independent biological experiments.
-
Figure 2—figure supplement 1—source data 1
PDF files containing original figures of immunoblot analysis displayed in Figure 2—figure supplement 1, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Original files of immunoblot analysis displayed in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig2-figsupp1-data2-v1.zip
-
Figure 2—figure supplement 1—source data 3
Numerical data used to generate Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig2-figsupp1-data3-v1.xlsx
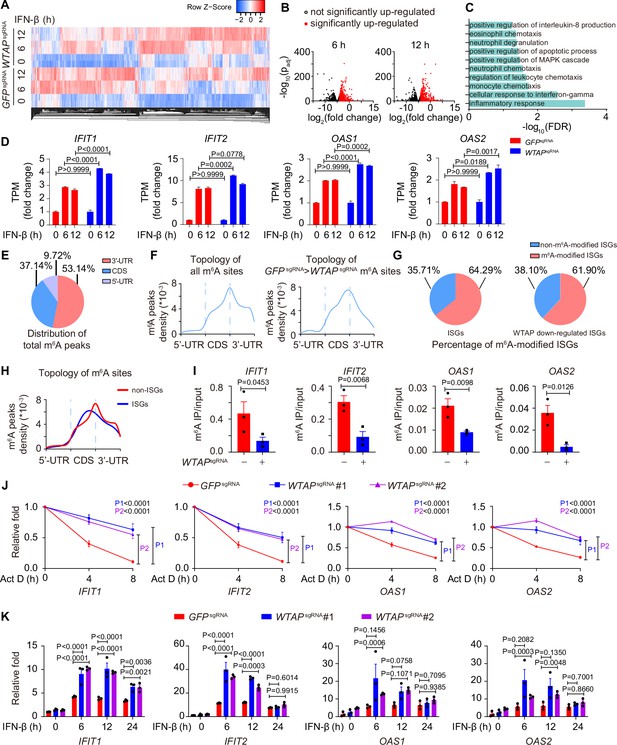
Wilm’s tumor-associated protein (WTAP) is crucial for the N6-methyladenosine (m6A) modification and expression of interferon-stimulated gene (ISG) mRNAs.
(A) Transcriptome sequencing analysis of control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages stimulated with 10 ng/mL IFN-β for 0, 6, 12 hr. The count-per-million (CPM) value of the ISGs was drawn by Heatmapper and clustered using the Centroid Linkage approach. (B) Volcano plots showing the changes in transcripts level of IFN-β up-regulated genes in WTAPsgRNA THP-1-derived macrophages versus control (GFPsgRNA) cells under IFN-β stimulation for 6 and 12 hr. Red dots indicated the significantly up-regulated genes in WTAPsgRNA cells (log2(fold change)>1 while adjusted p-value (padj) <0.05). (C) Gene ontology analysis for the WTAP down-regulated ISGs. (D) TPMs showing the changes in transcripts level of IFIT1, IFIT2, OAS1, and OAS2 in control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages stimulated with 10 ng/mL IFN-β for 6 or 12 hr or left untreated. (E, F) Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 4 hr, m6A modification analyzed by m6A methylated RNA immunoprecipitation (MeRIP) followed by deep sequencing (MeRIP-seq). Distribution (E) and topology (F) analysis (5’-untranslated regions (UTR), coding sequences (CDS) and 3’-UTR) of total m6A sites in control (GFPsgRNA) cells or WTAP-dependent m6A sites. (G) Percentage of m6A-modified ISGs including core ISGs or WTAP down-regulated ISGs were calculated and presented. (H) Topology analysis (5’-UTR, CDS, and 3’-UTR) of ISGs m6A sites and non-ISGs m6A sites. (I) Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 4 hr. MeRIP-quantitative real-time-polymerase chain reaction (qPCR) assay of IFIT1, IFIT2, OAS1, and OAS2 was performed and ratios between m6A-modified mRNA and input were shown (m6A IP/input). (J) Control (GFPsgRNA), WTAPsgRNA #1, and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 2 hr. After IFN-β treatment, medium with stimuli was replaced by 5 μM actinomycin D (Act D) for indicated time points. RNA was collected and detected by qPCR assay. (K) Control (GFPsgRNA), WTAPsgRNA #1, and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for indicated time points. Expression of IFIT1, IFIT2, OAS1, and OAS2 mRNA was analyzed through qPCR assays. All error bars, mean values ± SEM, p-values were determined by unpaired two-tailed Student’s t-test of n=3 independent biological experiments in (I). All error bars, mean values ± SEM, p-values were determined by a two-way ANOVA test of n=3 independent biological experiments in (J, K).
-
Figure 3—source data 1
Numerical data used to generate Figure 3.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig3-data1-v1.xlsx
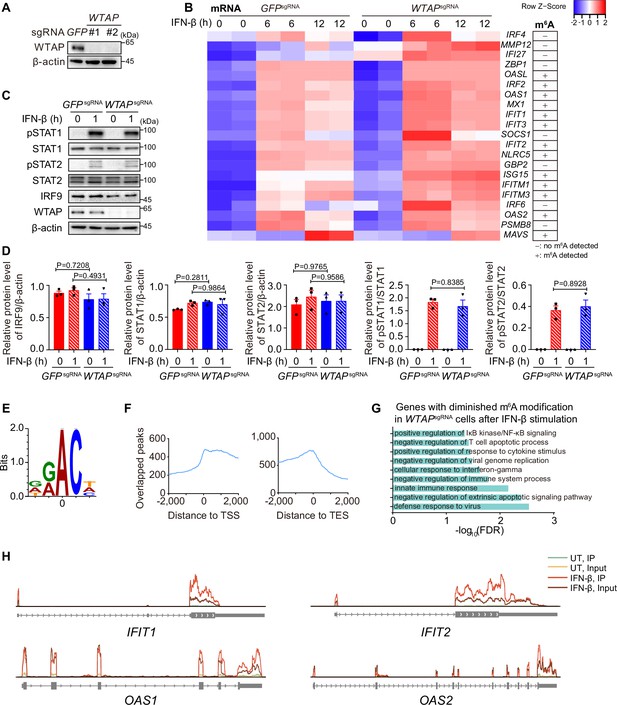
Wilm’s tumor-associated protein (WTAP) controls the N6-methyladenosine (m6A) modification of ISG mRNAs.
(A) WTAP knock-down efficiency of control (GFPsgRNA) and WTAPsgRNA #1 or #2 THP-1 was tested by immunoblot. (B) Transcriptome sequencing analysis of control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages stimulated with 10 ng/mL IFN-β for 0, 6, 12 hr, followed by RNA-seq. The count-per-million (CPM) value of core ISGs that upregulated in WTAPsgRNA THP-1 cells was drawn by Heatmapper and clustered using the Centroid Linkage approach. Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 4 hr, and m6A-modified ISGs analyzed by MeRIP-seq were shown. -: no m6A modification detected. +: m6A modification detected. (C, D) Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 1 hr or left untreated. Whole cell lysate (WCL) was collected, followed by immunoblot in (C), while relative protein level and phosphorylation level were measured and analyzed in (D). (E) Motif enrichment of m6A sites were analyzed in mRNAs of the IFN-β-induced WTAP-dependent m6A-modified genes through MeRIP-Seq. The position of the methylated adenosine is indicated as zero point. (F) Distance of m6A sites from translation start sites (TSS), translation end sites (TES) within the IFN-β-induced WTAP-dependent m6A modified genes were calculated and shown (zero point indicated position of the TSS or TES sites). (G) Gene ontology analysis for the WTAP down-regulated ISGs with lower m6A level in WTAPsgRNA #2 cells. (H) Distribution of m6A modification of IFIT1, IFIT2, OAS1, and OAS2. All error bars, mean values ± SD, p-values were determined by unpaired two-tailed Student’s t-test of n=3 independent biological experiments in (D). For (A, C), similar results were obtained for three independent biological experiments.
-
Figure 3—figure supplement 1—source data 1
PDF files containing original figures of immunoblot analysis displayed in Figure 3—figure supplement 1, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Original files of immunoblot analysis displayed in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig3-figsupp1-data2-v1.zip
-
Figure 3—figure supplement 1—source data 3
Numerical data used to generate Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig3-figsupp1-data3-v1.xlsx
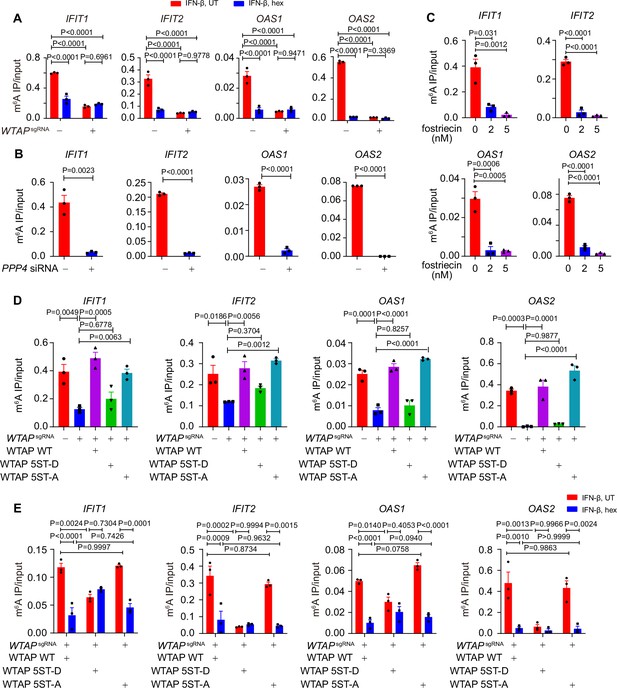
Liquid-phase separation of Wilm’s tumor-associated protein (WTAP) mediates N6-methyladenosine (m6A) modification and ISGs expression.
(A) Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β together with or without 5% 1,6-hexanediol (hex) and 20 μg/mL digitonin for 4 hr or left untreated. MeRIP-qPCR analysis of IFIT1, IFIT2, OAS1, and OAS2 was performed and ratios between m6A-modified mRNA and input were shown (m6A IP/input). (B) THP-1-derived macrophages transfected with scrambled (scr) siRNA or PPP4 siRNA were treated with 10 ng/mL IFN-β for 4 hr. MeRIP-qPCR assay of IFIT1, IFIT2, OAS1, and OAS2 was performed and ratios between m6A-modified mRNA and input were shown (m6A IP/input). (C) THP-1-derived macrophages were pre-treated with 2 nM or 5 nM fostriecin for 24 hr or left untreated, followed by 10 ng/mL IFN-β for 4 hr. MeRIP-qPCR analysis of IFIT1, IFIT2, OAS1, and OAS2 was performed and ratios between m6A-modified mRNA and input were shown (m6A IP/input). (D) Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages with or without expression of Flag-tagged wild-type (WT) WTAP, and its 5ST-D or 5ST-A mutants were treated with 10 ng/mL IFN-β for 4 hr. MeRIP-qPCR assay of IFIT1, IFIT2, OAS1, and OAS2 was performed and ratios between m6A-modified mRNA and input were shown (m6A IP/input). (E) Wild-type (WT) WTAP, 5ST-D or 5ST-A mutant-rescued WTAPsgRNA THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 4 hr. MeRIP-qPCR assay of IFIT1, IFIT2, OAS1, and OAS2 was performed and ratios between m6A-modified mRNA and input were shown (m6A IP/input). All error bars, mean values ± SEM, p-values were determined by two-way ANOVA test of n=3 independent biological experiments in (A, E). All error bars, mean values ± SEM, p-values were determined by unpaired two-tailed Student’s t-test of n=3 independent biological experiments in (B, C). All error bars, mean values ± SEM, p-values were determined by one-way ANOVA test of n=3 independent biological experiments in (D).
-
Figure 4—source data 1
Numerical data used to generate Figure 4.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig4-data1-v1.xlsx

Liquid-phase separation of Wilm’s tumor-associated protein (WTAP) is crucial for destabilization of ISG mRNAs.
(A, B) THP-1-derived macrophages were infected with Vesicular Stomatitis Virus (VSV) (m.o.i.=0.1) (A) or Herpes Simplex Virus-1 (HSV-1) (m.o.i.=1) (B) for indicated time points, along with or without 5% 1,6-hexanediol (hex) and 20 μg/mL digitonin for 2 hr. MeRIP-qPCR assay of IFIT1, IFIT2, OAS1 and OAS2 was performed and ratios between m6A-modified mRNA and input were shown (m6A IP/input). (C) Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β together with or without 5% hex and 20 μg/mL digitonin for 2 hr or left untreated. After IFN-β treatment, medium with stimuli was replaced by 5 μM actinomycin D (Act D) for indicated time points. RNA was collected and detected by qPCR assay. (D) Control (GFPsgRNA) and WTAPsgRNA THP-1-derived macrophages were transfected with scrambled (scr) or PPP4-targeted siRNA. Cells were treated with 10 ng/mL IFN-β for indicated time points. Expressions of IFIT1 and IFIT2 mRNA were detected by qPCR assay. (E) THP-1-derived macrophages were pre-treated with 2 nM or 5 nM fostriecin for 24 hr or left untreated, followed by 10 ng/mL IFN-β for indicated time points. Expression of IFIT1, IFIT2, OAS1, and OAS2 mRNA was analyzed through qPCR assays. (F) Immunoblot analysis of the expression of wild-type (WT) WTAP,5ST-D or 5ST-A mutant in WT WTAP, 5ST-D, or 5ST-A mutant-rescued WTAPsgRNA THP-1-derived macrophages. All error bars, mean values ± SEM, p-values were determined by two-way ANOVA test of n=3 independent biological experiments in (A–E). For (F), similar results were obtained for three independent biological experiments.
-
Figure 4—figure supplement 1—source data 1
PDF files containing original figures of immunoblot analysis displayed in Figure 4—figure supplement 1, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original files of immunoblot analysis displayed in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig4-figsupp1-data2-v1.zip
-
Figure 4—figure supplement 1—source data 3
Numerical data used to generate Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig4-figsupp1-data3-v1.xlsx
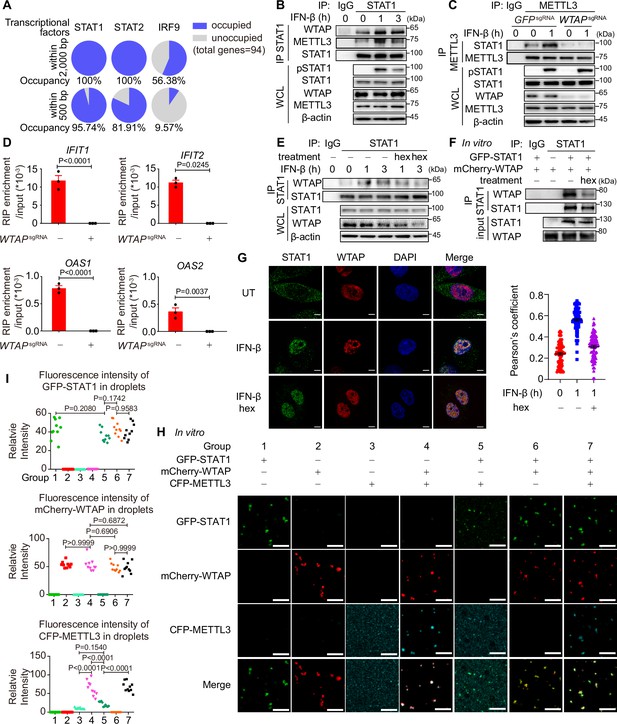
Liquid-phase separated Wilm’s tumor-associated protein (WTAP) bridges STAT1 and N6-methyladenosine (m6A) modification methyltransferase complex to direct m6A modification on ISG mRNAs.
(A) Occupancy of the promoter of the WTAP down-regulated ISGs with lower m6A level in WTAPsgRNA #2 cells (genes were identified in Figure 3—figure supplement 1G and both –2000–0 bp and –500–0 bp upstream of transcription start site were analyzed) by STAT1, STAT2, and IRF9 was predicted by AnimalTFDB 3.0. (B) THP-1-derived macrophages were treated with 10 ng/mL IFN-β for indicated time points. Whole cell lysate (WCL) was collected and immunoprecipitation (IP) experiment using anti-STAT1 antibody or rabbit IgG was performed, followed by immunoblot. (C) Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 1 hr or left untreated. WCL was collected and IP experiment using anti-METTL3 antibody or rabbit IgG was performed, followed by immunoblot. (D) Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 2 hr or left untreated. RNA immunoprecipitation (RIP) experiments using anti-METTL3 antibody or rabbit IgG control were performed in control and WTAPsgRNA #2 THP-1-derived macrophages treated with 10 ng/mL IFN-β for 2 hr. RNA of ISGs enriched by RIP was analyzed by quantitative real time-polymerase chain reaction (qPCR) assay, and the ratios between RIP-enriched RNA and input were shown (RIP enrichment/input). (E) THP-1-derived macrophages were treated with 10 ng/mL IFN-β for indicated time points together with or without 5% 1,6-hexanediol (hex) and 20 μg/mL digitonin. WCL was collected and IP experiment with anti-STAT1 antibody or rabbit IgG was performed, followed by immunoblot. (F) Recombinant GFP-STAT1 (10 μM) and mCherry-WTAP (10 μM) were incubated with or without 5% hex in vitro. IP experiment with anti-STAT1 antibody or rabbit IgG was performed, followed by immunoblot. (G) THP-1-derived macrophages were treated with 10 ng/mL IFN-β only or with 5% hex and 20 μg/mL digitonin for 1 hr or left untreated (UT). Interaction between STAT1 (green) and WTAP (red) were imaged using confocal microscope. Pearson’s correlation coefficient was analyzed and calculated from n=80 cells through ImageJ. Scale bars indicated 5 μm. (H, I) Recombinant GFP-STAT1 (10 μM), mCherry-WTAP (10 μM), and CFP-METTL3 (10 μM) were incubated using physiological buffer at 37℃ in vitro. After incubation, images were captured using a confocal microscope (H). Relative fluorescence intensity of proteins in n=10 condensates were analyzed by ImageJ software (I). All error bars, mean values ± SEM, p-values were determined by unpaired two-tailed Student’s t-test of n=3 independent biological experiments in (D). All error bars, mean values ± SD, p-values were determined by unpaired two-tailed Student’s t-test of n=80 cells in (G) and n=3 independent biological experiments in (I). For (B–D, E–G, H), similar results were obtained for three independent biological experiments.
-
Figure 5—source data 1
PDF files containing original figures of immunoblot analysis displayed in Figure 5, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig5-data1-v1.zip
-
Figure 5—source data 2
Original files of immunoblot analysis displayed in Figure 5.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig5-data2-v1.zip
-
Figure 5—source data 3
Numerical data used to generate Figure 5.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig5-data3-v1.xlsx
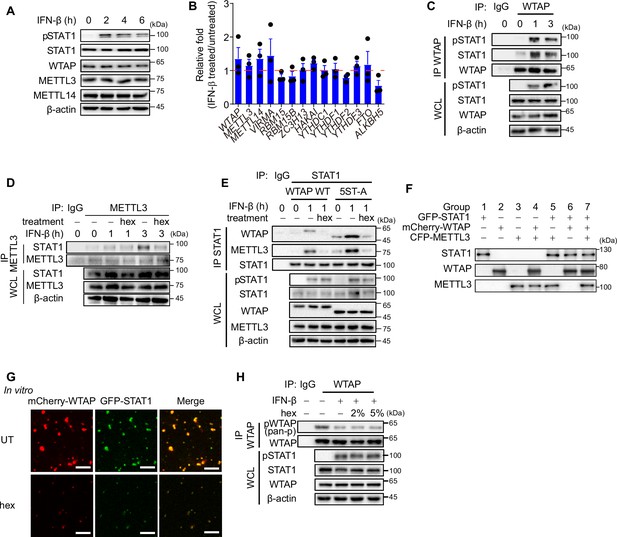
Liquid-phase separated Wilm’s tumor-associated protein (WTAP) promotes the interaction between STAT1 and METTL3.
(A) THP-1-derived macrophages were treated with 10 ng/mL IFN-β for indicated time points. Expression of indicated proteins were detected through immunoblot. (B) THP-1-derived macrophages were treated with 10 ng/mL IFN-β for indicated time points. Expressions of components of m6A methyltransferase complex, m6A readers, and m6A demethylase mRNA were detected through quantitative real-time polymerase chain reaction (qPCR) assay. (C) THP-1-derived macrophages were treated with 10 ng/mL IFN-β for indicated time points. Whole cell lysate (WCL) was collected and immunoprecipitation (IP) experiment using anti-WTAP antibody or rabbit IgG was performed, followed by immunoblot. (D) THP-1-derived macrophages were treated with 10 ng/mL IFN-β for indicated time points together with or without 5% 1,6-hexanediol (hex) and 20 μg/mL digitonin. WCL was collected and IP experiment with anti-METTL3 antibody or rabbit IgG was performed, followed by immunoblot. (E) Wild-type (WT) WTAP or 5ST-A mutant-rescued WTAPsgRNA THP-1-derived macrophages are stimulated with 10 ng/mL IFN-β together with or without 5% hex and 20 μg/mL digitonin for 1 hr or left untreated. WCL was collected and IP experiment using anti-STAT1 antibody or rabbit IgG was performed, followed by immunoblot. (F) Protein level of different recombinant proteins of different groups in Figure 5H and I was detected through immunoblot assays. (G) Recombinant GFP-STAT1 (10 μM) and mCherry-WTAP (10 μM) were incubated with 5% hex or left untreated (UT) in 37℃. Interaction between WTAP and STAT1 was imaged using a confocal microscope. Scale bars indicated 10 μm. (H) THP-1-derived macrophages were treated with 10 ng/mL IFN-β together with or without 2% or 5% hex and 20 μg/mL digitonin for 1 hr or left untreated. WCL was collected and IP experiment using anti-WTAP antibody or rabbit IgG was performed, followed by immunoblot. pWTAP was detected by anti-phosphoserine/threonine/tyrosine antibody (pan-p). All error bars, mean values ± SEM, p-values were determined by unpaired two-tailed Student’s t-test of n=3 independent biological experiments in (B). For (A, C–H), similar results were obtained for three independent biological experiments.
-
Figure 5—figure supplement 1—source data 1
PDF files containing original figures of immunoblot analysis displayed in Figure 5—figure supplement 1, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Original files of immunoblot analysis displayed in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig5-figsupp1-data2-v1.zip
-
Figure 5—figure supplement 1—source data 3
Numerical data used to generate Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig5-figsupp1-data3-v1.xlsx
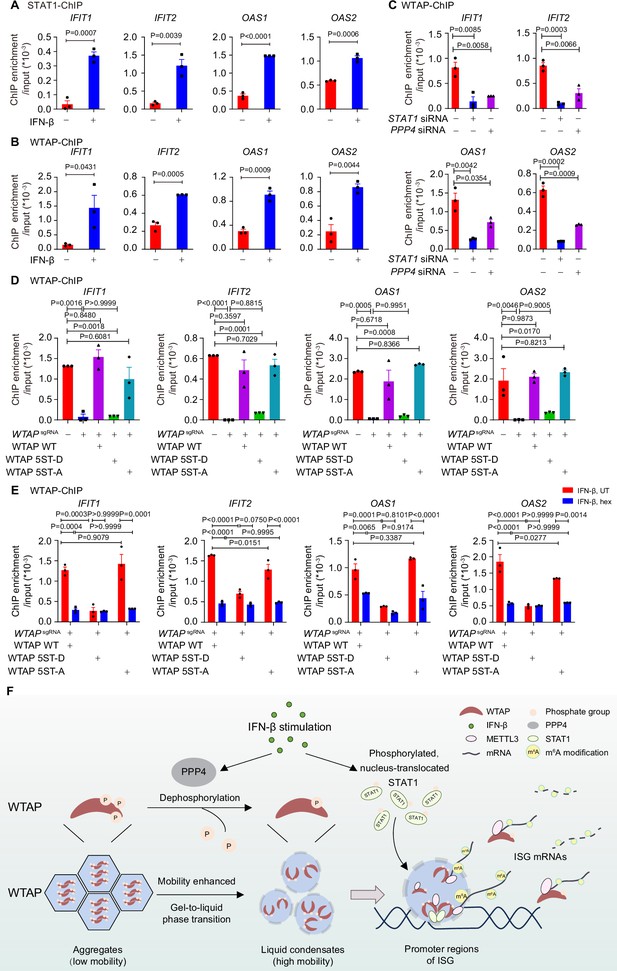
Phase transition of Wilm’s tumor-associated protein (WTAP) mediates its interaction with ISG promoter regions to regulate N6-methyladenosine (m6A) modification on ISG mRNAs.
(A) Chromatin immunoprecipitation (ChIP) experiments using anti-STAT1 antibody or rabbit IgG control were performed in THP-1-derived macrophages treated with 10 ng/mL IFN-β for 2 hr or left untreated. Binding between the promoter regions of IFIT1, IFIT2, OAS1, and OAS2 with WTAP was detected by quantitative real-time polymerase chain reaction (qPCR) assay. Ratios between ChIP-enriched DNA and input were shown (ChIP enrichment/input). (B) ChIP experiments using anti-WTAP antibody or rabbit IgG control were performed in THP-1-derived macrophages treated with 10 ng/mL IFN-β for 2 hr or left untreated. Binding between the promoters of IFIT1, IFIT2, OAS1, and OAS2 with WTAP were detected by qPCR assay. Ratios between ChIP-enriched DNA and input were shown (ChIP enrichment/input). (C) ChIP experiments using anti-WTAP antibody or rabbit IgG control were performed in THP-1-derived macrophages transfected with scrambled (scr) siRNA, STAT1 siRNA, or PPP4 siRNA treated with 10 ng/mL IFN-β for 2 hr. Binding between the promoter regions of IFIT1, IFIT2, OAS1, and OAS2 with WTAP was detected by qPCR assay. Ratios between ChIP-enriched DNA and input were shown (ChIP enrichment/input). (D) ChIP experiments using anti-WTAP antibody or rabbit IgG control were performed in control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages with or without expression of Flag-tagged wild-type (WT) WTAP, and its 5ST-D or 5ST-A mutants treated with 10 ng/mL IFN-β for 2 hr. Binding between the promoter regions of IFIT1, IFIT2, OAS1, and OAS2 with WTAP was detected by qPCR assay. Ratios between ChIP-enriched DNA and input were shown (ChIP enrichment/input). (E) ChIP experiments using anti-WTAP antibody or rabbit IgG control were performed were performed in WT WTAP, 5ST-D, or 5ST-A mutant-rescued WTAPsgRNA THP-1-derived macrophages treated with 10 ng/mL IFN-β along with 5% 1,6-hexanediol (hex) for 2 hr. Binding between the promoters of IFIT1, IFIT2, OAS1, and OAS2 with WTAP were detected by qPCR assay. Ratios between ChIP-enriched DNA and input were shown (ChIP enrichment/input). (F) Schematic figure of IFN-β-induced phase transition of WTAP regulates the m6A modification of ISG mRNAs. All error bars, mean values ± SEM, p-values were determined by unpaired two-tailed Student’s t-test of n=3 independent biological experiments in (A–C). All error bars, mean values ± SEM, p-values were determined by one-way ANOVA test of n=3 independent biological experiments in (D). All error bars, mean values ± SEM, p-values were determined by two-way ANOVA test of n=3 independent biological experiments in (E).
-
Figure 6—source data 1
Numerical data used to generate Figure 6.
- https://cdn.elifesciences.org/articles/100601/elife-100601-fig6-data1-v1.xlsx
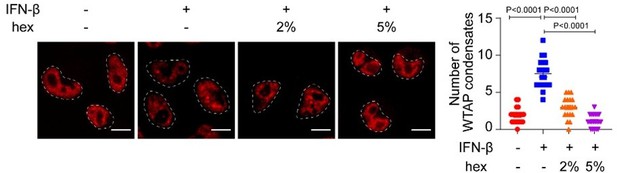
mCherry-WTAP-rescued HeLa cells were treated with 10 ng/mL IFN-β together with or without 2% or 5% hex and 20 μg/mL digitonin for 1 hr or left untreated.
Phase separation of mCherry-WTAP was observed through confocal microscopy. The number of WTAP condensates that diameter over 0.4 μm of n = 20 cells were counted through ImageJ and shown. Scale bars indicated 10 μm. All error bars, mean values ± SD, P-values were determined by unpaired two-tailed Student’s t-test of n = 20 cells in (B). For (A), similar results were obtained for three independent biological experiments.

Wild type (WT) WTAP or 5ST-A mutant-rescued WTAPsgRNA THP-1-derived macrophages are stimulated with or without with 10 ng/mL IFN-β together followed by 2% or 5% 1,6-hexanediol (hex) and 20 μg/mL digitonin for 1 hr or left untreated.
Antibody and imaged using confocal microscope. Scale bars indicated 10 μm.
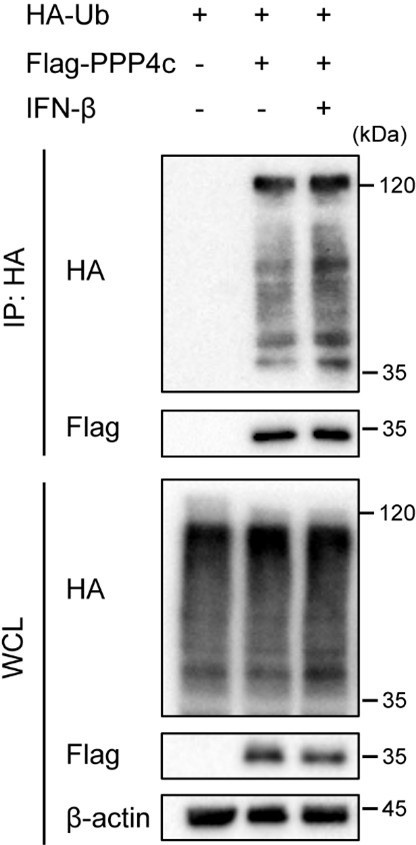
HEK 293T transfected with HA-ubiquitin (HA-Ub) and Flag-PPP4 were treated with 10 ng/mL IFN-β or left untreated.
Whole cell lysate (WCL) was collected and immunoprecipitation (IP) experiment using anti-Flag antibody was performed, followed by immunoblot. Similar results were obtained for three independent biological experiments.
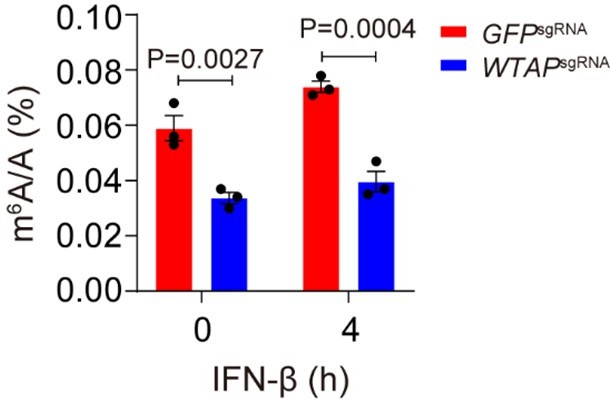
Control (GFPsgRNA) and WTAPsgRNA #2 THP-1-derived macrophages were treated with 10 ng/mL IFN-β for 4 hours.
Global m6A level was detected and quantified through ELISA assays. All error bars, mean values ± SEM, P-values were determined by two-way ANOVA test independent biological experiments.
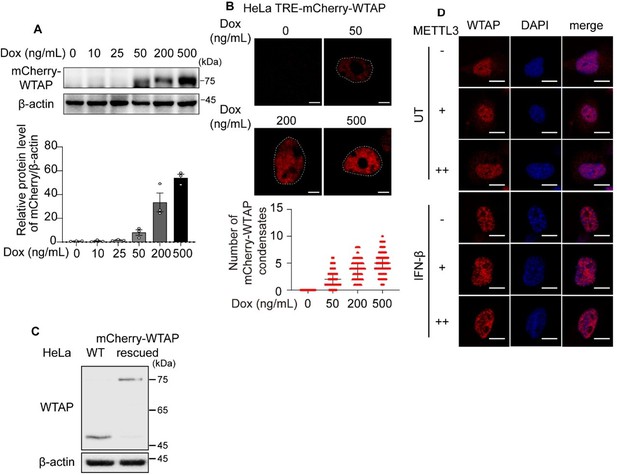
(A) Immunoblot analysis of the expression of mCherry-WTAP in TRE-mCherry-WTAP HeLa cells treated with different doses of doxycycline (Dox). Protein level of mCherry-WTAP was quantified and normalized to β-actin of n=3 independent biological experiments. Results were obtained for three independent biological experiments. (B) Phase separation diagram of mCherry-WTAP in TRE-mCherry-WTAP HeLa cells treated with different doses of Dox were observed through confocal microscopy. Representative images for three independent biological experiments were shown in b while number of WTAP condensates that diameter over 0.4 μm of n=80 cells were counted and shown as medium with interquartile range. Dotted white lines indicated the location of nucleus. Scale bars indicated 10 μm. (C) Immunoblot analysis of the expression of endogenous WTAP in wildtype (WT) HeLa cells and mCherry-WTAP-rescued WTAPsgRNA HeLa cells. (D) mCherry-WTAP-rescued HeLa cells were transfected with 0, 200 or 400 ng of Flag-METTL3, followed with 10 ng/mL IFN-β for 1 hour or left untreated (UT). Phase separation of mCherry-WTAP was observed through confocal microscopy. The number of WTAP condensates that diameter over 0.4 μm of n = 20 cells were counted through ImageJ and shown. Representative images of n=20 cells were shown. All error bars, mean values ± SD were determined by unpaired two-tailed Student’s t-test of n = 3 independent biological experiments in (A). For (A, C), similar results were obtained for three independent biological experiments.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Goat anti-mouse HRP | Invitrogen | Cat#A-16072; RRID:AB_2534745 | IB (1:4000) |
| Antibody | Goat anti-rabbit HRP | Invitrogen | Cat#A-16104; RRID:AB_2534776 | IB (1:4000) |
| Antibody | Anti-rabbit IgG | Beyotime Biotechnology | Cat#A7016; RRID:AB_2905533 | IP (1 μg) |
| Antibody | Anti-mouse IgG | Beyotime Biotechnology | Cat#A7028; RRID:AB_2909433 | IP (1 μg) |
| Antibody | Goat anti-rabbit IgG (Alexa Fluor 488) | Invitrogen | Cat#A-11034; RRID:AB_2576217 | IF (1:500) |
| Antibody | Goat anti-mouse IgG (Alexa Fluor 568) | Invitrogen | Cat#A-11031; RRID:AB_144696 | IF (1:500) |
| Antibody | Anti-mCherry | Proteintech | Cat#26765–1-AP; RRID:AB_2876881 | IB (1:3000) |
| Antibody | Anti-β-actin | Sigma-Aldrich | Cat#A1978; RRID:AB_476692 | IB (1:5000) |
| Antibody | Anti-Flag (M2) | Sigma-Aldrich | Cat#A8592; RRID:AB_439702 | IB (1:3000) |
| Antibody | Anti-WTAP | Santa Cruz Biotechnology | Cat#sc-365500; RRID:AB_10843970 | IF (1:150) |
| Antibody | Anti-WTAP | Bethyl Laboratories | Cat#14994 | IB (1:1000) IP (1:100) IF (1:100) |
| Antibody | Anti-m6A | Synaptic Systems | Cat#202003; RRID:AB_2279214 | IP (1:400) |
| Antibody | Anti-phosphoserine/threonine/tyrosine (pan-p) | Invitrogen | Cat#61–8300; RRID:AB_138456 | IB (1:1000) |
| Antibody | Anti-phospho-STAT1 (Tyr701) | Cell Signaling Technology | Cat#9167; RRID:AB_561284 | IB (1:1000) |
| Antibody | Anti-STAT1 | Cell Signaling Technology | Cat#14994; RRID:AB_2737027 | IB (1:1000) IF (1:150) IP (1:100) |
| Antibody | Anti-phospho-STAT2 (Tyr690) | Cell Signaling Technology | Cat#88410; RRID:AB_2800123 | IB (1:1000) |
| Antibody | Anti-STAT2 | Cell Signaling Technology | Cat#72604; RRID:AB_2799824 | IB (1:1000) |
| Antibody | Anti-IRF9 | Cell Signaling Technology | Cat#76684; RRID:AB_2799885 | IB (1:1000) |
| Antibody | Anti-METTL3 | Proteintech | Cat#15073–1-AP; RRID:AB_2142033 | IB (1:1000) IF (1:150) |
| Antibody | Anti-PPP4 | Proteintech | Cat#10262–1-AP; RRID:AB_2300020 | IB (1:1000) |
| Chemical compound, drug | protein A/G Agarose | Pierce | Cat#20333, Cat#20399 | IP |
| Chemical compound, drug | anti-Flag M2 affinity gel | Sigma-Aldrich | Cat#A2220 | IP |
| Cell line (Homo sapiens) | THP-1 | Cell Bank of the Chinese Academy of Sciences (Shanghai, China) | RRID:CVCL_0006 CSTR:19375.09.3101HUMSCSP567 | Cultured in RPMI 1640 with 10% FBS and 1% L-glutamine |
| Cell line (H. sapiens) | HEK 293T | Cell Bank of the Chinese Academy of Sciences (Shanghai, China) | RRID:CVCL_0063 CSTR:19375.09.3101HUMSCSP5209 | Cultured in DMEM with 10% FBS and 1% L-glutamine |
| Cell line (H. sapiens) | HeLa | Cell Bank of the Chinese Academy of Sciences (Shanghai, China) | RRID:CVCL_0030 CSTR:19375.09.3101HUMSCSP504 | Cultured in DMEM with 10% FBS and 1% L-glutamine |
| Chemical compound, drug | Human IFN-β recombinant protein | PeproTech Inc | Cat#300-02BC | Cytokines stimulation |
| Chemical compound, drug | phorbol-12-myristate-13-acetate (PMA) | Sigma-Aldrich | Cat#P1585 | THP-1 differentiation (100 nM) |
| Chemical compound, drug | Puromycin | Sigma-Aldrich | Cat#P9620 | 1–10 μg/mL |
| Chemical compound, drug | 1,6-hexanediol | Sigma-Aldrich | Cat#240117 | Phase separation inhibitor (5%) |
| Chemical compound, drug | Actinomycin D | Sigma-Aldrich | Cat#SBR00013 | RNA synthesis inhibitor (5 μM) |
| Chemical compound, drug | Digitonin | Sigma-Aldrich | Cat#D141 | Phase separation experiments (20 μg/mL) |
| Chemical compound, drug | DTT | Sigma-Aldrich | Cat#3483-12-3 | Protein purification (1 mM) |
| Chemical compound, drug | DAPI | Sigma-Aldrich | Cat#D9542 | IF (1 μg/mL) |
| Chemical compound, drug | NP-40 | Beyotime Biotechnology | Cat#P0013F | Protein purification (0.1%) |
| Chemical compound, drug | Fostriecin | Abcam | Cat#ab144255 | PPP4 inhibitor (2–5 nM) |
| Chemical compound, drug | LipoRNAiMAX | Invitrogen | Cat#13778100 | siRNA transfection reagent |
| Chemical compound, drug | Isopropyl-beta-D-thiogalactopyranoside (IPTG) | MIKX | Cat#CA413 | Recombinant Protein expression (1 mM) |
| Chemical compound, drug | Superluminal High- efficiency Transfection Reagent | MIKX | Cat#11231804 | Lentiviral Plasmids transfection reagent |
| Chemical compound, drug | Hieff-Trans Liposomal Transfection Reagent | Yeasen | Cat#40802ES02 | Plasmid transfection reagent |
| Chemical compound, drug | TRIzol reagent | Invitrogen | Cat#15596026 | RNA extraction |
| Chemical compound, drug | Dynabeads mRNA Purification Kit | Invitrogen | Cat#61006 | mRNA purification |
| Chemical compound, drug | HiScript III RT SuperMix for qPCR (+gDNA wiper) | Vazyme | Cat#R323-01 | RNA reverse-transcription |
| Chemical compound, drug | 2×PolarSignal SYBR Green mix Taq | MIKX | Cat#MKG900-10 | RT-qPCR |
| Sequence-based reagent (H. sapiens) | WTAP sgRNA #1 | This paper | sgRNA targeting WTAP for CRISPR-Cas9 system | Sequence: GCTGTAGTCCTGCTGGTACT |
| Sequence-based reagent (H. sapiens) | WTAP sgRNA #2 | This paper | sgRNA targeting WTAP for CRISPR-Cas9 system | Sequence: AAGTTGTGCAATACGTCCCT |
| Sequence-based reagent (H. sapiens) | GFP sgRNA | This paper | sgRNA targeting GFP as control for CRISPR-Cas9 system | Sequence: CATGCCGAGAGTGATCCCGG |
| Sequence-based reagent (H. sapiens) | PPP4 siRNA | This paper | siRNA targeting PPP4 | Sequence: CGGCUACCUAUUUGGCAGUGA |
| Sequence-based reagent (H. sapiens) | STAT1 siRNA | This paper | siRNA targeting STAT1 | Sequence: GAAAGAGCUUGACAGUAAA |
| Software, algorithm | ImageJ | National Institutes of Health | RRID:SCR_003070 | Image analysis (Version 1.52) |
| Software, algorithm | GraphPad Prism | GraphPad Software | RRID:SCR_002798 | Statistical analysis (Version 8.0) |
| Software, algorithm | Leica Application Suite X | Leica microsystems | RRID:SCR_013673 | Image analysis (Version 4.2) |
Additional files
-
Supplementary file 1
Primers for qPCR, RIP-qPCR, MeRIP-qPCR, and ChIP-qPCR.
- https://cdn.elifesciences.org/articles/100601/elife-100601-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/100601/elife-100601-mdarchecklist1-v1.docx





