Mechanistic insight for T-cell exclusion by cancer-associated fibroblasts in human lung cancer
Figures
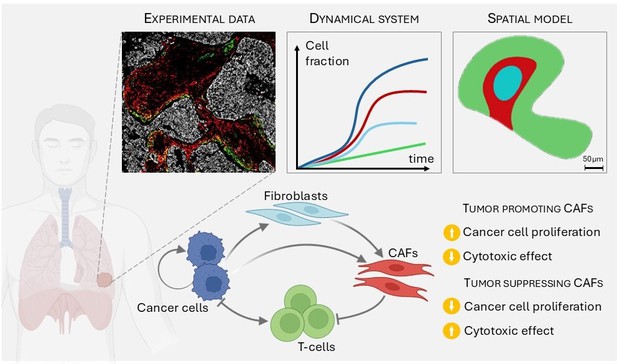
Summary of the article: Starting from experimental data on the structure and density of the different species on lung cancer, we first build a dynamical system, and then a spatiotemporal model.
We incorporate four species with specific interactions, that may lead to different outcomes.
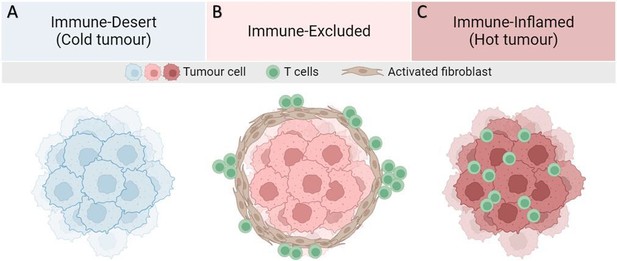
Major T-cell infiltration patterns observed in solid tumors.
(A) Lack of tumor antigen, inadequate priming, defects in antigen presentation and/or lack of presentation, and/or lack of T-cell-attracting chemokines result in the absence of T-cells in the tumor. (B) Presence of T-cells in invasive margins but absent in the tumor bed. Immune evasion may be due to stromal barriers, lack of chemokines, aberrant vasculature, or hypoxia. (C) High degree of T-cell infiltration forms a hot tumor.
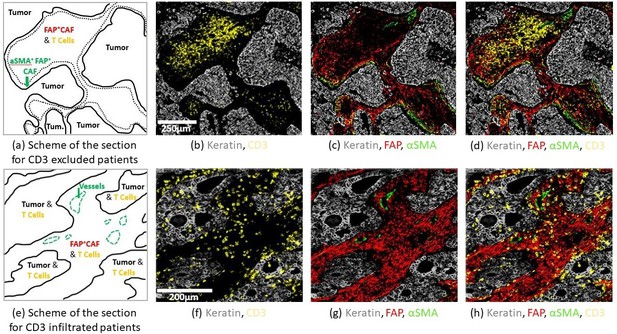
Structural organization in human Non Small Cell Lung Cancer (NSCLC).
Staining was performed by multiplex immuno-histochemistry (IHC). FFPE NSCLC sections were stained for keratin as a marker of cancer cells (gray), CD3 (yellow), and fibroblast markers αSMA (green), FAP (red). First row: CD3 excluded patient. (a) Scheme of the section showing CD3+ cell exclusion from tumor nests. The green arrow highlights border regions with contractile fibroblast barrier αSMA+ FAP+ and low CD3+ cells. (b) CD3+ cells are localized in the center of the stroma. (c) Dense αSMA staining at the tumor border are associated with a decrease of CD3+ cell abundance. (d) Recap of all the markers. Second row: CD3 infiltrated patient. (e) Scheme of the section showing CD3+ infiltration in the tumor islets. The green arrow shows αSMA+ staining on vessels. (f) CD3+ are localized in the tumor nest. (g) FAP+ staining is localized throughout the stroma. (h) Recap of all the markers.
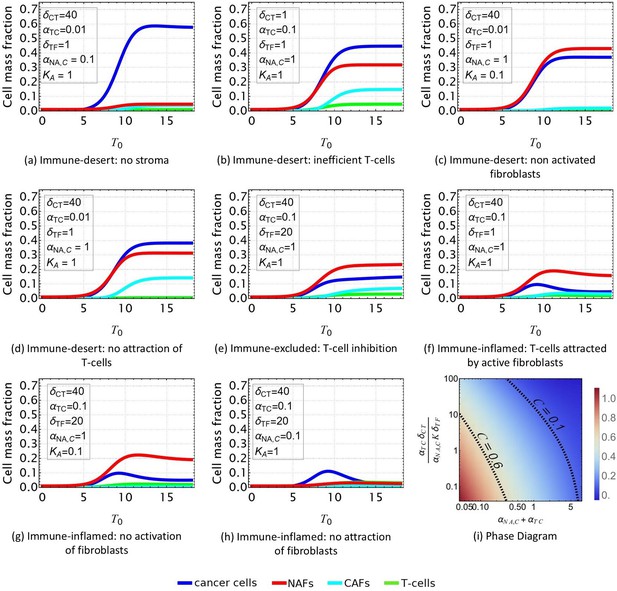
Evolution of the tumor microenvironment (TME) composition over time.
Different profiles are obtained from Equations 1–4 according to the set of parameters reported in Table 2 and consistent with the scenarios shown in Figure 2. In all the plots, and . (a, b, c, d) Immune-desert tumor. Cancer cells proliferate when the immune response is inefficient or when T-cells are not attracted to the tumor. e: Immune-excluded tumor. The cancer-associated fibroblast (CAF) barrier inhibits T-cell infiltration, and promotes tumor growth. (f, g, h) Immune-inflamed tumor. T-cell infiltration limits cancer cell growth. (i) Density plot of the equilibrium cancer cell fraction at fixed and, in function of the two parameters that control the growth: reflects the ability of T-cells to kill the tumor, and reflects the competition between species for space and resources.
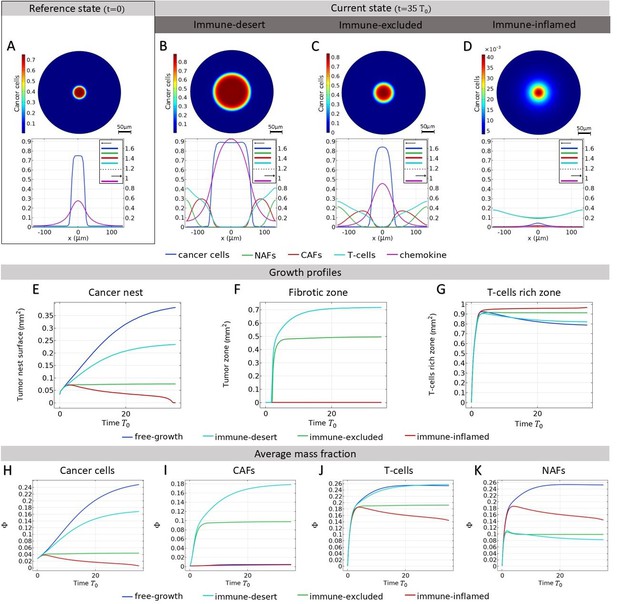
Small solid tumor growth.
We refer here to the different tumor phenotypes described in Figure 2. (A) Mass fraction of cancer cells at time and profile of different mass fractions on a section of the tumor. (B-C-D) Mass fraction of cancer cells at time and profile of mass fraction in immunodeficient tumor (B), when immune infiltration is inhibited by cancer-associated fibroblasts (CAFs) (C) and in immune-inflamed tumor (D) In D, the scale of the colorbar is 10−3 the values of (A), (B), (C), since this panel represents the case of efficient T-cells. (E-F-G) Development of different zones. (E) Surface fraction profile of the tumor nest for different scenarios, calculated as. (F) Fibrotic surface fraction profile for different scenarios, calculated as. (G) T-cell rich area fraction profile for different scenarios, calculated as . (H) Cancer cell average mass fractions. (I) CAF average mass fractions. (J) T-cell average mass fractions. (K) Non-activated fibroblast (NAF) average mass fractions.
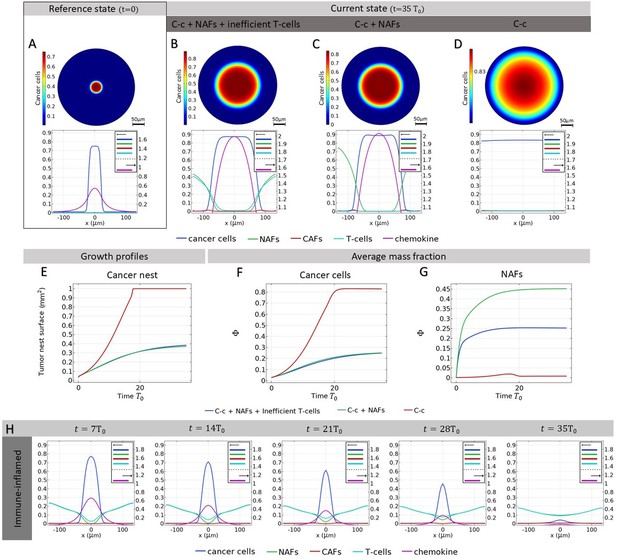
Free growth of a tumor nest.
(A-G) Cancer population density plots for different scenarios: Cancer cells (C-c) + non-activated fibroblasts (NAFs) + Inefficient T-cells, Cancer cells + NAFs,Cancer cells alone. (A) Mass fraction of cancer cells at time and profile of different mass fractions on a section of the tumor. (B) Mass fraction of cancer cells at time and mass fraction profile for a case of a stroma composed by C-c, NAFs, and inefficient T-cells. (C) Mass fraction of cancer cells at time and profile of the different mass fractions on a cut of the tumor for a case of a stroma composed by C-c and NAFs. (D) Mass fraction of cancer cells at time and mass fraction profile for a case with no stroma in the tumor microenvironment. (E) Surface fraction of the tumor nest in the different scenarios. (F) Average mass fraction of the cancer cells in the different scenarios. (G) Average mass fraction of the NAFs in the different scenarios. (H) Time evolution of the mass fraction profile of the cut of the simulation window in the case of a tumor in the presence of efficient T-cells and NAFs only, without T-cells.
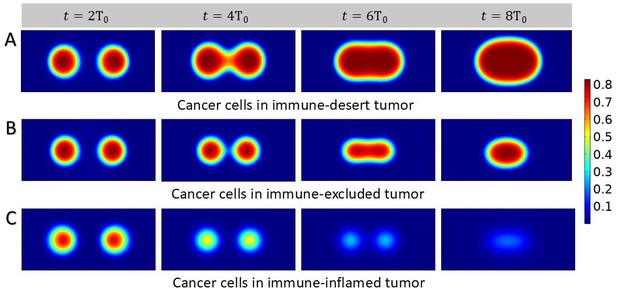
Growth process of two small solid tumors.
Density plots showing cancer cell population. We start with two tumor nests placed at the same distance from the center of the domain (30 μm), sharing the same mass fraction of cancer cells and we present different scenarios. (A) Nests coalesce in immune-desert tumor, i.e., in the presence of inefficient T-cells. (B) Nests interacting in immune-excluded tumor, with chemotactic T-cells and cancer-associated fibroblasts (CAFs). (C) Nests coalesce in immune-inflamed tumor.
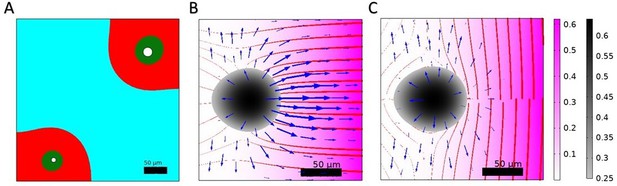
Anisotropy.
Following biological observation reported in Figure 3, we here show different cases for describing anisotropy. (A) Example of the tumor microenvironment in the presence of two blood vessels (in white). The tumor nest is in cyan (), the cancerous stroma in red () and the healthy stroma in green (). The tumor shape is dictated by the localization of the blood vessels from which T-cells and non-activated fibroblasts (NAFs) are issued. (B-C) Tumor behavior in the presence of normal (B) or orthonormal (C) fibers. The tumor fraction is indicated in colors from black to gray which we limit for to better visualize of the contour of the nest. The flux is indicated with blue arrows whose thickness is proportional to the magnitude of the flux. The nematic order is shown with red lines whose thickness is proportional to the determinant of the matrix (see SI, Nematic order) and the fibroblast fraction is indicated with a pink color gradient.
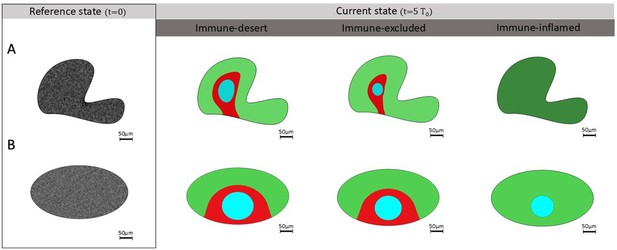
Different tumor phenotypes in non-spherical domains.
Deformed shapes result in anisotropic configurations by forcing non-activated fibroblasts (NAFs) and T-cells to enter the domain from only one side of the boundary. Different colors indicate separated regions where the nest (in light blue) is confined by the boundary of cancer-associated fibroblasts (CAFs) (in red) within a healthy stroma (in green). (A) Immune-inflamed case leads to disappearance cancer cells at . (B) Elliptical area leads to slower dynamics.
Tables
Composition of the tumor microenvironment.
This table presents the literature about the fraction of different species in the lung TME. The values found in the present studies are written in the two last rows (raw data is provided as Supplementary Material). Different methods have been used, in different subtypes of lung cancer. We introduce the following abbreviations. (C-c): Cancer cells. (MΦ): Macrophages. (T-c): T-cells. (Fb): Fibroblasts. (scRNA-seq): single-cell RNA seq,(NSCLC): Non Small Cell Lung Cancer, (LUAD): Adenocarcinoma, (SSN): Sub-Solid Nodule. (S): stroma region. (T): tumor nest region. (ST): Stroma + tumor nest region. (TSR): tumor stroma ratio. Data for Ref (Ireland et al., 2020; Kim et al., 2020; Laughney et al., 2020; Maynard et al., 2020; Qian et al., 2020) were extracted from Curated Cancer Cell Atlas, 2023.
| Method | Sample | C-c (%) | T-c (%) | Fb (%) | MΦ(%) | TSR |
|---|---|---|---|---|---|---|
| Staining Sieren et al., 2010 | LUAD | >25 | >20 | 0.3 | ||
| Staining Sieren et al., 2011 | LUAD | 67 | >6.5 | 2.6 | ||
| scRNA-seq Lambrechts et al., 2018 | NSCLC | 55 (S) | 4 (S) | 15 (S) | ||
| scRNA-seq Ireland et al., 2020 | SCLC | 76 | ||||
| scRNA-seq Kim et al., 2020 | LUAD | 33 | 19.5 | ∼6 | 20.4 | |
| scRNA-seq Laughney et al., 2020 | LUAD | <12 | 42.6 | ∼1 | 12.2 | |
| scRNA-seq Maynard et al., 2020 | NSCLC | <22.7 | 16 | 8 | 23 | |
| scRNA-seq Qian et al., 2020 | mixed | 19.9 | 28.1 | ∼2 | 27.5 | |
| scRNA-seq Xing et al., 2021 | LUAD | 16.4 | 30 | ∼2 | 18.4 | |
| scRNA-seq Altorki et al., 2022 | LUAD | 12 | 7.5 | |||
| Staining Mathieson et al., 2022 | NSCLC | 75 (S), 0 (T) | ||||
| Staining (present article) | LUAD | 47±13 | 31±17 (S), 0 (T) | 1.07±0.68 | ||
| Staining (present article) | LUSC | 54±10 | 35±12 (S), 0(T) | 1.30±0.53 |
-
Table 1—source data 1
Raw data for the two last lines of the table.
- https://cdn.elifesciences.org/articles/101885/elife-101885-table1-data1-v1.xlsx
Scaling variation and estimation of the coefficients according to different scenarios.
The scenarios are described in Section Model Parameters and Fixed Point Analysis and shown in Figure 4. The coefficients above are those introduced in Equations 1–4. The left column summarizes the different roles that T-cells can play in a cell mixture and the values of the coefficients of the mixture are listed in the following horizontal lines of the table. The scaling of is always: 1 , and .
| Control of: | Killing C-c by T-c | Attraction of T-c by C-c | Inhibition of T-c by F-c | Attraction of F-c to C-c | Activation of F-c |
| Efficient T-cells (T-c) but no attraction by (C-c) | No role | No role | No role | ||
| Efficient T-cells but inhibited by (F-c) | 1∼1 | ||||
| Inefficient T-cellsno need of fibroblasts | No role | No role | No role | ||
| Efficient T-cells not inhibited by fibroblasts | 1∼1 | ||||
| Efficient T-cells and fibroblasts not attracted | |||||
| Efficient T-cells fibroblasts not activated |
Values of the parameters in the spatial model.
| Parameter | Value | Parameter | Value |
|---|---|---|---|
Parameter values varying depending on the scenario in Figure 5—figure supplement 1.
| Parameter | ||
|---|---|---|
| C-c+NAFs+inefficient T-cells | 2.5×10−3 | 2.5×10−3 |
| C-c+NAFs | 2.5×10−6 | 2.5×10−3 |
| C-c | 2.5×10−6 | 2.5×10−3 |





