Expression signature based on TP53 target genes doesn't predict response to TP53-MDM2 inhibitor in wild type TP53 tumors
Figures
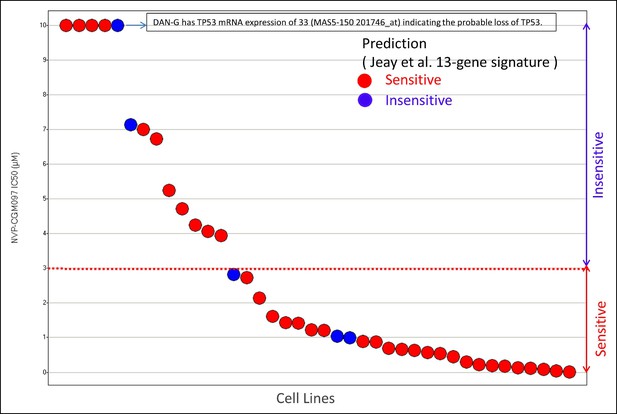
Cell lines sensitivity to NVP-CGM097 in validation set of 40 likely TP53 WT cell lines.
https://doi.org/10.7554/eLife.10279.005Tables
List of 12 cell lines with inactivated TP53 in the validation set of 52 cancer cell lines considered to be TP53 wild-type by Jeay et al., (2015).
| Cell line name | TP53 inactivating mutation(s) | Alternative reads/reference reads | TP53 mRNA (MAS5-150 201746_at) | TP53 CN ratio | Jeay et al. 13-gene signature prediction | NVP-CGM097 sensitivity |
|---|---|---|---|---|---|---|
| KASUMI-1 | p. R248Q | 52/0 | 265 | 0.54 | insensitive | insensitive |
| COLO-818 | p. C135R | 34/0 | 257 | 1.14 | insensitive | insensitive |
| IGR-37 | p. C229fs | 110/11 | 9 | 0.59 | insensitive | insensitive |
| HCC202 | p. T284fs | 35/4 | 14 | 0.8 | insensitive | insensitive |
| EFM-192A | p. F270fs | 7/1 | 10 | 0.74 | insensitive | insensitive |
| NCI-H1568 | p. H179R | 89/1 | 202 | 0.82 | insensitive | insensitive |
| COLO-783 | p. P27L | 38/0 | 304 | 1.05 | sensitive | insensitive |
| GA-10 | p. I232N, p. P152L | 94/50, 52/76 | 493 | 0.81 | insensitive | insensitive |
| VMRC-RCW | p. I332_splice | 192/68 | 63 | 1.65 | insensitive | insensitive |
| JHH-5 | p. PPQH190del | 107/41 | 272 | 1.03 | insensitive | insensitive |
| HDLM-2 | 1 | 0.94 | insensitive | insensitive | ||
| RERF-LC-KJ | 25 | 1.3 | insensitive | insensitive |
Performance of Jeay et al., (2015) 13-gene signature prediction in validation set of 40 likely TP53 wild-type cancer cell lines.
| Performance measure | Cell sensitivity defined by NVP-CGM097 |
|---|---|
| Sensitivity | 89% (24/27) |
| Specificity | 15% (2/13) {DAN-G removal 8% (1/12) *} |
| PPV | 68.6% (24/35) |
| NPV | 40% (2/5) {DAN-G removal 25% (1/4) *} |
| Response rate | 67.5% (27/40) |
-
* DAN-G has TP53 mRNA expression of 33 (MAS5-150 201746_at) indicating the probable loss of TP53 mRNA.
-
(Stringent TP53 mRNA expression cutoff is set at 32 (MAS5-150 201746_at) to indicate loss of TP53 mRNA).
-
NPV - negative predicted value; PPV - positive predicted value.
Additional files
-
Supplementary file 1
(A) 40 likely TP53 WT cancer cell lines from Jeay et al. (2015) validation set. (BPerformance of Jeay et al. (2015) 13-gene Signature Prediction in validation set of 28 likely TP53 WT cancer cell lines with sensitivity defined by NVP-CFC218. (NVP-CFC218 data was available for 28 out of 40 likely TP53 WT cancer cell lines.) (CList of 29 cell lines with inactivated TP53 in the set of 113 cancer cell lines considered to be TP53 wild type by Jeay et al. (2015). (D) 84 likely TP53 WT cancer cell lines from Jeay et al. (2015). (E) COSMIC information on mutations in cell lines listed in the Table 1.
- https://doi.org/10.7554/eLife.10279.006





