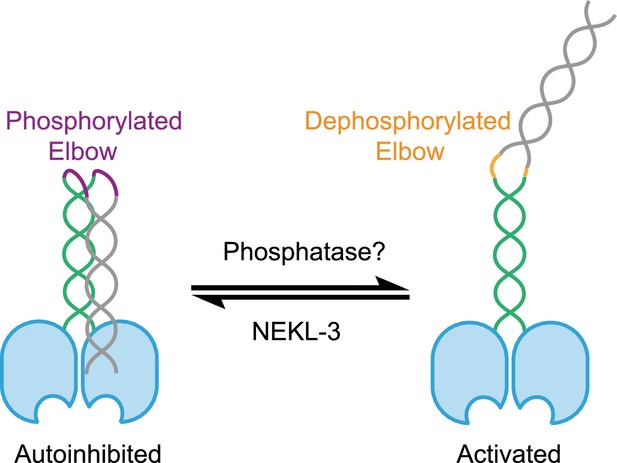
Kinesin-2 autoinhibition requires elbow phosphorylation
Figures
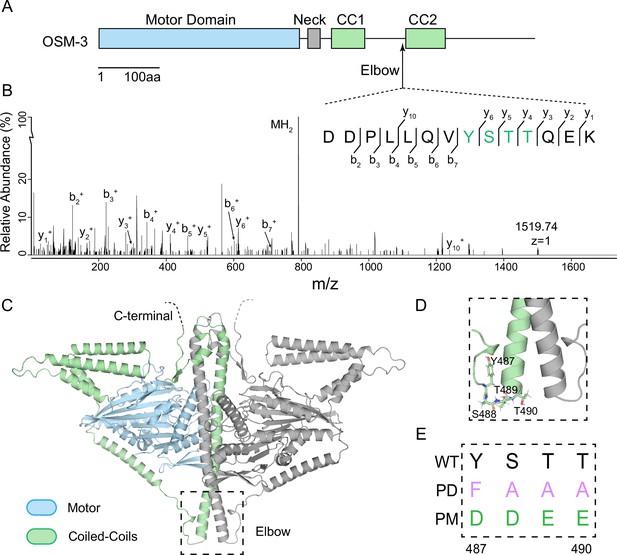
NEKL-3 phosphorylates OSM-3 at its ‘elbow’.
(A) Schematic of the full-length OSM-3. Motor domain (blue), neck (gray), and coiled-coils (green) are indicated. CC, coiled-coil. (B) Mass spectrum of an OSM-3 peptide that was phosphorylated by NEKL-3. Phosphorylated gel bands were subjected to MS analysis searching for phosphorylation modifications. Residues 487–490 of OSM-3 were phosphorylated and were marked by green color. (C) Phosphorylated residues 488–490 are at the ‘elbow’ of OSM-3. It shows the overall structure of the homodimeric OSM-3 predicted by AlphaFold2. The dashed square marks the ‘elbow’ region and is zoomed-in in D. (E) Genome-editing constructs of the elbow, showing the PD (phosphor-dead) and PM (phosphor-mimic) sequences comparing to wild type. Residues 487–490 of OSM-3 were edited to ‘FAAA’ for PD strain or edited to ‘DDEE’ for PM strain. Abbreviations: Y, Tyr; S, Ser; T, Thr; F, Phe; A, Ala; D, Asp; E, Glu.
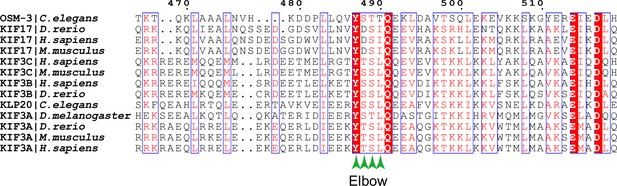
Kinesin-2 family members have a conserved elbow motif.
Sequence alignment of OSM-3 and other members in the kinesin-2 family (KIF3A, KIF3B, KIF3C, and KIF17) from different model organisms as indicated. This figure shows the alignment results around the elbow region. Green arrowheads indicate the four phosphorylated residues. The position of amino acids of OSM-3 is labeled at the top of the figure. Sequences were aligned using CLUSTAL O (1.2.4) and presented by ESPript 3.0.

Predicted aligned error (PAE) and predicted Local Distance Difference Test (pLDDT) of OSM-3 dimer models.
(A) PAE of OSM-3 dimer presented by LocalColabFold. A total of five predicted models were presented, and the ‘rank_1’ model was selected for the following analysis. x-axis, residue number; y-axis, A and B represent two identical OSM-3 peptide chains. (B) pLDDT for all five predicted models shown in (A).
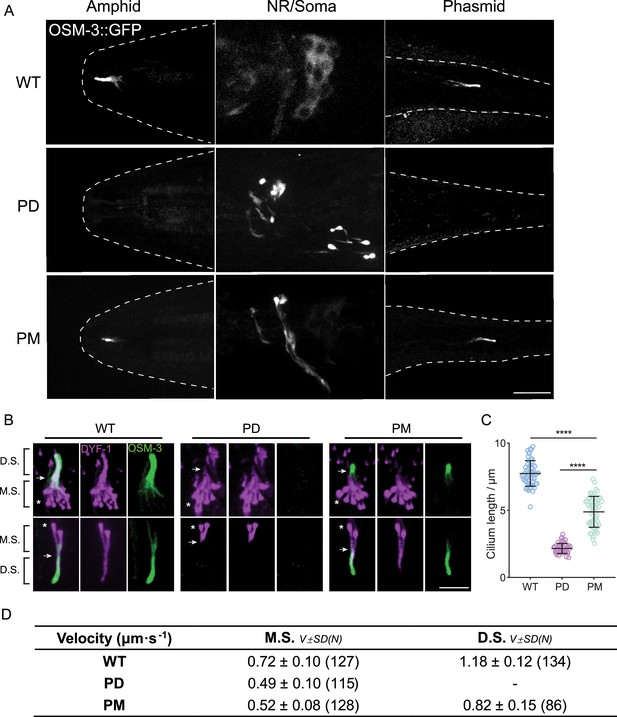
Phosphorylation at the elbow of OSM-3 is inhibitive in vivo.
(A) Representative images of the phospho-dead (PD) and phospho-mimic (PM) knock-in worms showing their OSM-3 signal at amphid cilia, amphid neuronal soma, and phasmid cilia, respectively. The contours of the worms are marked by white dashed lines. Scale bar, 10 μm. (B) Representative images of the cilia from PD and PM worms marked by the ciliary marker DYF-1::mScarlet. White arrows indicate the junction between middle and distal segments, while the asterisks indicate the ciliary base. M.S., middle segment; D.S., distal segment. Scale bar, 5 μm. (C) Statistics of the cilium length of the strains shown in (B). The lengths of DYF-1::mScarlet signals were measured and analyzed. (D) Intraflagellar transport (IFT) velocities of PD and PM worms. ****p < 0.0001, analyzed by one-way ANOVA, p values were adjusted by BH method.
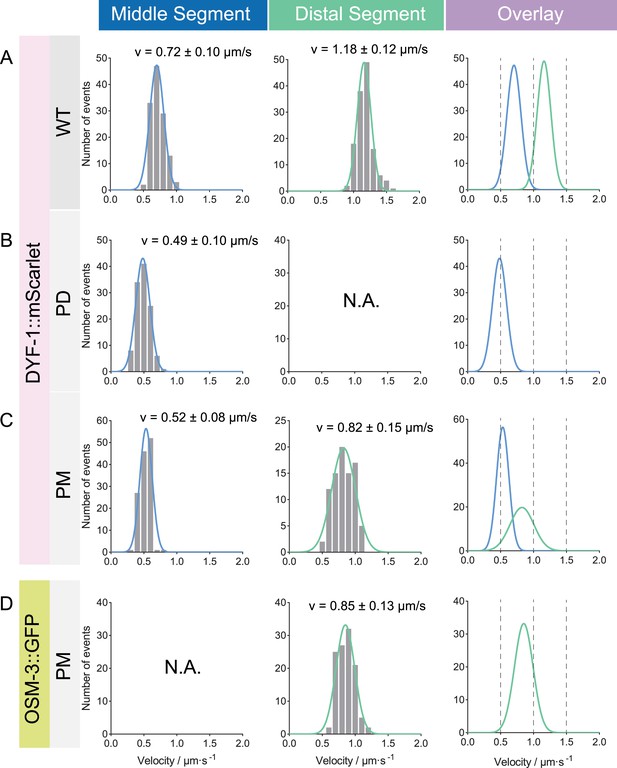
Statistical analysis of the intraflagellar transport (IFT) velocities in osm-3pd and osm-3pm worms, corresponding to Figure 2D.
(A–C) IFT velocity marked by DYF-1-mScarlet. Genotypes are shown on the left. Left panel, frequency distribution of IFT particles in the middle segment. Middle panel, frequency distribution of IFT particles in the distal segment. Data were fitted with a Gaussian distribution. Right panel, overlay of the fitted curves of middle and distal segments. (D) OSM-3::GFP velocity in osm-3pm worms. N.A., not available.
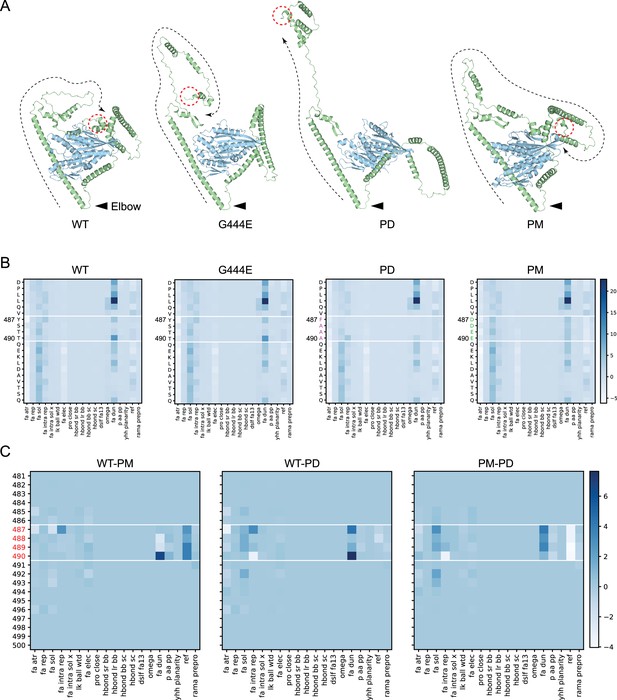
Structural models of the OSM-3 kinesin and its mutants.
(A) Relaxed structure models of OSM-3 and mutants. Black arrowheads indicate the elbow while red dashed circles mark the C-terminus of the protein. Black dashed lines showed the extending direction from the elbow toward the C-terminus. Wild-type (WT) and phosphor-mimic (PM) showed close interaction between the tail and motor domain, while G444E and phosphor-dead (PD) showed that the tails are far away from the motor. (B) Heatmaps of the energy states of the pre-relaxed structure models from amino acid 481st to 500th, as labeled on the left; the amino acids between the white lines are the elbow region; each row represents an energy item as labeled on the bottom. (C) Heatmaps comparing the energy states by direct subtraction between the mutants and WT. The PD mutant has lower ‘fa_dun’ energy while having higher ‘ref’ energy than that of the PM mutant. Energy terms are explained in Figure 3—source data 1.
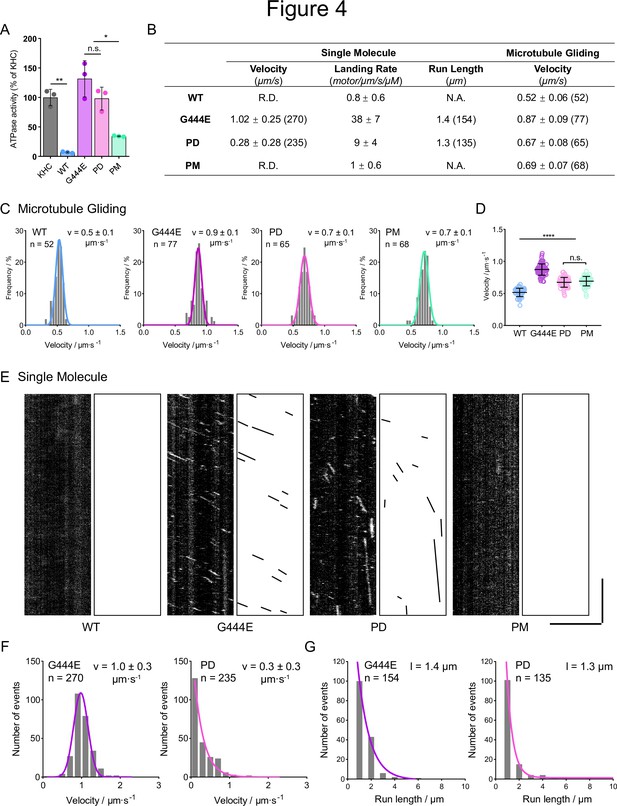
Phospho-dead OSM-3 behaves constitutively active in vitro while phospho-mimic OSM-3 stays autoinhibited.
(A) Microtubule-stimulated ATPase activity of wild-type (WT) OSM-3 and mutants. G444E, the hyperactive positive control; KHC, kinesin heavy chain. Average activity of KHC was set to 100% and others were normalized to KHC. (B) Summary of the single-molecular assay and the microtubule gliding assay. R.D., rarely detected. N.A., not available. Data are [mean ± SD (number of events)]. (C) Velocity distributions of microtubule gliding assays of the indicated OSM-3 constructs. n, total events measured. v, μm s–1, average velocity with standard deviation. (D) Statistics of microtubule gliding velocities shown in (C). (E) Representative kymographs of the single-molecular movements of WT OSM-3 and mutants as indicated. Scale bars, vertical, 10 s; horizontal, 5 μm. (F) Velocity distributions of the single-molecular assays. n, total events measured. v, μm s–1, average velocity with standard deviation. The distribution of G444E was fitted with a Gaussian distribution curve, while the distribution of phosphor-dead (PD) was fitted with a one-phase decay curve. (G) Run length distributions of the single-molecular assays. n, total events measured. l, average run length. The curves were fitted with the one-phase decay distribution. *p < 0.05, **p < 0.01, ****p < 0.0001, analyzed by one-way ANOVA, p values were adjusted by BH method.
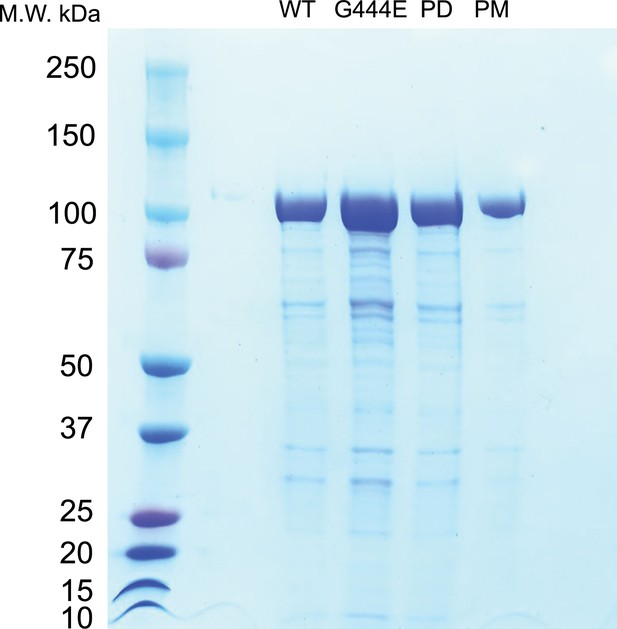
SDS–PAGE of purified recombinant OSM-3 mutants.
SDS–PAGE of the purified recombinant OSM-3::eGFP and mutants, Coomassie Blue stained. The elution peaks of each recombinant protein were shown, labeled on the top.
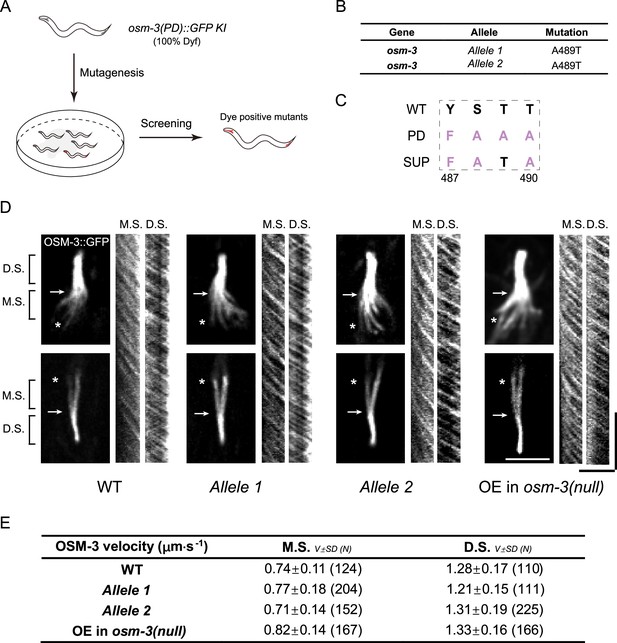
Genetic screening identified T489 as the key regulatory residue in the elbow of OSM-3.
(A) Schematics of the forward genetic screen. 100% Dyf osm-3(PD)::GFP KI worms were mutated by ethyl methanesulfonate (EMS) and F2 progenies were screened for dye filling positive mutants. (B) Two independent suppressor mutants cloned from the genetic screening. (C) Amino acid sequences of the suppressor mutants at the elbow. (D) Representative images of the cilia of the suppressors and the kymographs showing the velocity of OSM-3. The rightmost panel shows the same OSM-3 version (487–489: ‘FATA’) with the suppressors but over-expressed under the ciliary Pdyf-1 promoter in osm-3(p802) worm. The arrows indicate the junction between middle and distal segments, while the asterisks indicate the ciliary base. M.S., middle segment; D.S., distal segment. Scale bars, vertical, 10 s; horizontal, 5 μm. (E) Summary of the OSM-3 velocity. M.S., middle segment. D.S. distal segment. Data are [mean ± SD (number of events)].
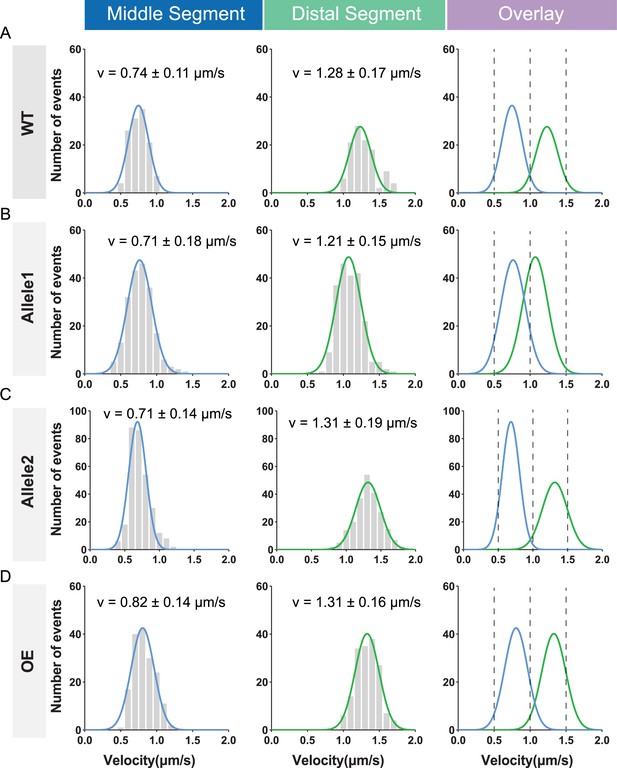
Statistical analysis of the intraflagellar transport (IFT) velocities in osm-3pd suppressors, corresponding to Figure 5E.
(A–C) OSM-3 velocity of wild-type (WT) and mutant proteins marked by GFP. Genotypes are shown on the left. Left panel, frequency distribution of IFT particles in the middle segment. Middle panel, frequency distribution of IFT particles in the distal segment. Data were fitted with the Gaussian distribution. Right panel, overlay of the fitted curves of middle and distal segments. (D) Velocity and frequency distribution of overexpressed OSM-3 (487–489: ‘FATA’) under the ciliary Pdyf-1 promoter in osm-3(p802) worm.
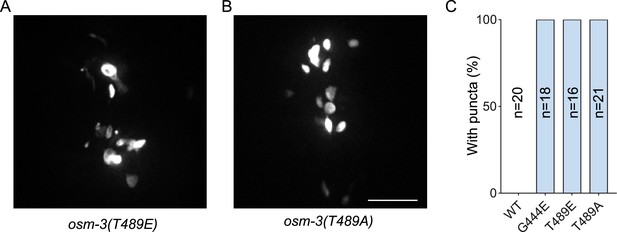
T489E and T489A of OSM-3 cause aggregated signals.
(A, B) Representative images of OSM-3(T489E) and OSM-3(T489A) overexpressed in osm-3(p802) strain under the ciliary Pdyf-1 promoter, respectively. The images showed the areas around nerve rings. Scale bar, 10 μm. (C) Statistics of strains in (A, B), showing the percentage of worms with bright puncta around nerve rings for each strain. n, number of worms checked.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/103648/elife-103648-mdarchecklist1-v1.pdf
-
Supplementary file 1
Mass spectrometry results of NEKL-3 treated OSM-3.
- https://cdn.elifesciences.org/articles/103648/elife-103648-supp1-v1.xlsx
-
Supplementary file 2
Energy minimization results of models showed in Figure 3.
- https://cdn.elifesciences.org/articles/103648/elife-103648-supp2-v1.xlsx
-
Supplementary file 3
WGS results of the strain GOU5380.
- https://cdn.elifesciences.org/articles/103648/elife-103648-supp3-v1.xlsx
-
Supplementary file 4
WGS results of the strain GOU5381.
- https://cdn.elifesciences.org/articles/103648/elife-103648-supp4-v1.xlsx
-
Supplementary file 5
C. elegans strains used in this study.
- https://cdn.elifesciences.org/articles/103648/elife-103648-supp5-v1.docx