Axial contraction and short-range compaction of chromatin synergistically promote mitotic chromosome condensation
Figures
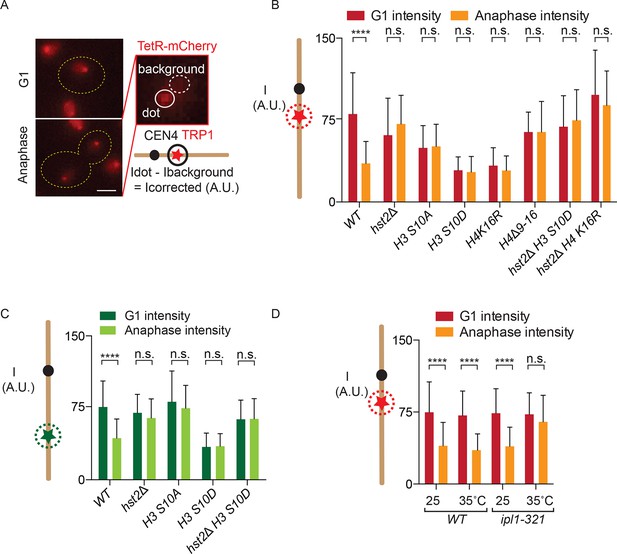
Fluorescence intensity of TetO/TetR-mCherry as a read-out for chromatin compaction.
(A) Representative images of a cell in G1 and anaphase, containing a TetO array at the TRP1 locus and expressing TetR-mCherry (red). Fluorescence intensity of a focus is measured by determining the total fluorescence and subtracting the background, giving the corrected fluorescence intensity. Scale bar is 2 µm. (B) TetR-mCherry intensities for the indicated wild type (WT) and mutant strains in G1 and anaphase mother cells. One way Analysis Of Variance ( ANOVA) was performed to test significance. (C) Fluorescence intensity for a wild type strain containing LYS4:LacO and expressing LacI-GFP. Student’s t-test was performed to determine significance. (D) Anaphase TetR-mCherry intensities for the indicated strains, synchronized in G1 by alpha-factor treatment and released at the indicated temperatures. Intensities for G1 were determined 5 min after release from alpha-factor induced arrest. All data are means and standard deviation for n>30 cells. **** p<0.0001 and n.s. not significant.
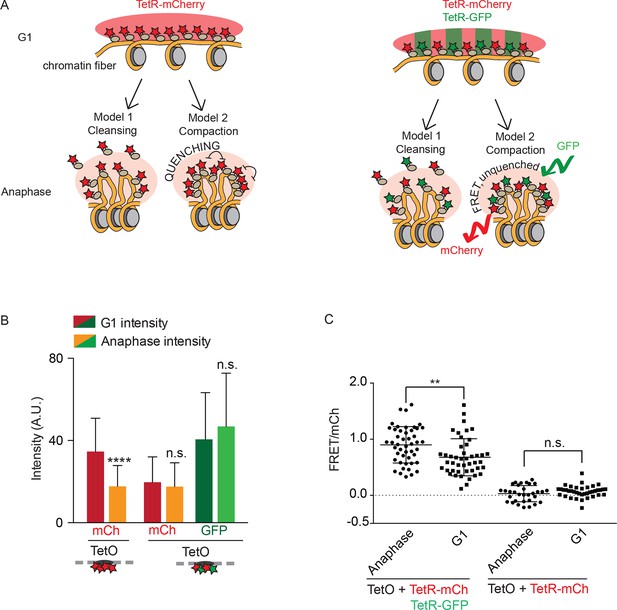
Fluorophore quenching causes changes of TetO/TetR intensity over the cell cycle.
(A) Two models to explain anaphase-specific decrease in fluorescence brightness (cleansing and quenching, see text for explanations). Shown are the consequences of each model in G1 and anaphase, in the case of cells carrying TRP1:TetOs and either expressing only TetR-mCherry or TetR-mCherry and TetR-GFP. (B) G1 and anaphase TetO/TetR-mCherry intensities in cells carrying TetO and expressing only TetR-mCherry (left) or TetR-mCherry and TetR-GFP (right). Data are means and standard deviations, unpaired Student’s t tests were performed to test significance, **** p<0.0001 and n.s. not significant. (C) FRET values for indicated strains. Plotted are mean values and standard deviation. Unpaired Student’s t tests were performed to test significance, ** p<0.01 and n.s. not significant.
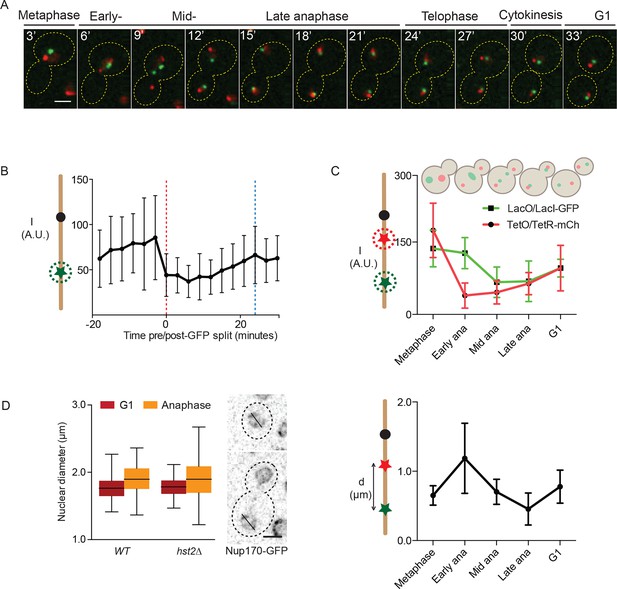
Dynamics of chromatin compaction and chromosome arm contraction.
(A) Example of a cell going from metaphase to the next G1 phase with TRP1 and LYS4 loci marked with TetR-mCherry and LacI-GFP, respectively. (B) Background normalized, mean GFP-intensity values of mitotic cells, aligned at mid-anaphase (red dashed line: GFP dot split). Blue line indicates formation of the first bud. Standard deviations are shown. (C) Upper panel: normalized (to G1) intensity of TetR-mCherry and LacI-GFP foci in mother cells in the indicated cell cycle stages. Lower panel: mother TRP1:TetO - LYS4:LacO distances in indicated cell cycle stages. Shown are mean and standard deviation for n>30 cells. (D) Nuclear diameter of G1 and late anaphase cells in wild type and hst2∆ cells containing Nup170-GFP. Box shows median value, whiskers all data points n>50 cells. Scale bars are 2 µm.
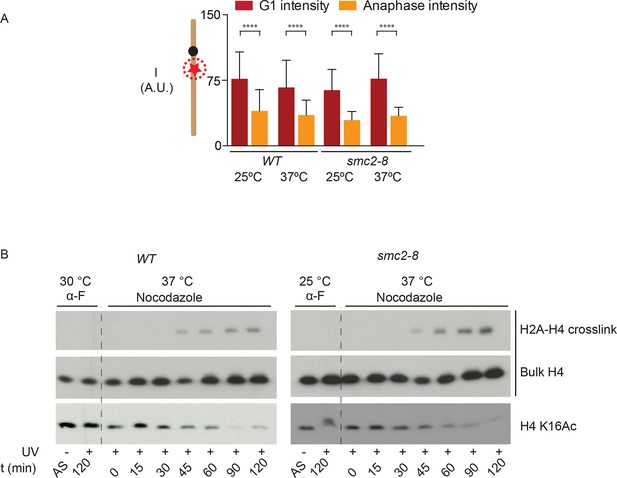
Condensin does not impact chromatin compaction.
(A) TetR-mCherry intensities in the mother cell for the indicated strains and cell cycle stages. To inactivate Smc2, cells were shifted to 37ºC for 90 min. One way ANOVA was performed to test significance, **** p<0.0001 and n>40. (B) Yeast cells producing H2A Y58BPA were synchronized with alpha-factor at permissive temperature and then released into medium containing nocodazole at restrictive temperature. Samples were taken at indicated times, irradiated with UV and histones extracted with acid from isolated nuclei. Western blot against H4 detects the H2A-H4 crosslink (upper row), bulk H4 (lower row) and blotting against H4 K16Ac shows cell cycle progression.
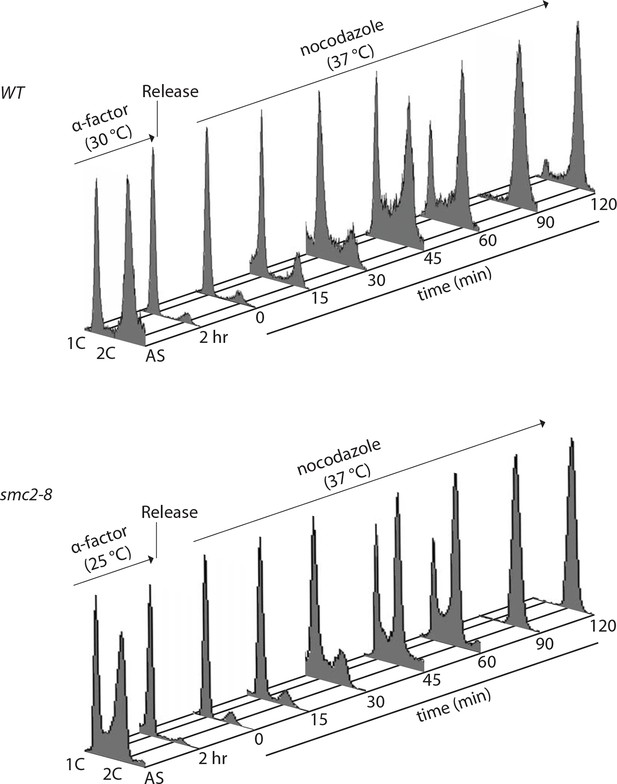
FACS analysis of alpha-factor synchronized cells released into nocodazole (wild type or smc2-8).
The same samples were analysed by western blot in Figure 4B.
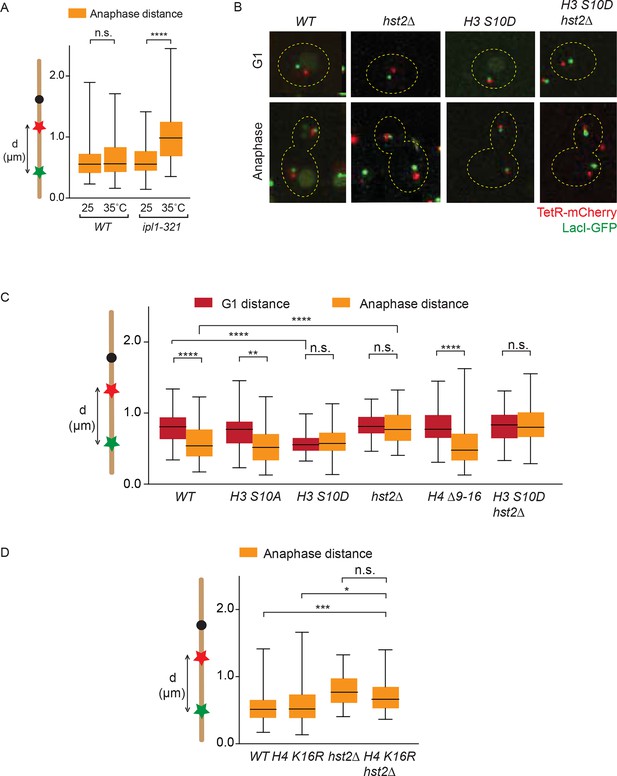
Chromatin compaction does not influence axial chromosome contraction.
(A) TRP1-LYS4 distances for the indicated strains, synchronized in G1 by alpha-factor treatment and released at the indicated temperatures. (B) TRP1-LYS4 distances were determined in the mother cell for the indicated strains and cell cycle stages. Box shows median value, whiskers all data points n>30 cells. One way ANOVA was performed to test significances, ** p<0.01, **** p<0.0001, n.s. not significant. (C) Example cells containing the indicated mutations and their impact on chromosome length as determined by the TRP1 (red) to LYS4 (green) distance. (D) TRP1-LYS4 distances were determined for the indicated strains in anaphase. Box shows median value, whiskers all data points n>45. One way ANOVA was performed to test significances, *** p<0.001, * p<0.05, n.s. not significant.
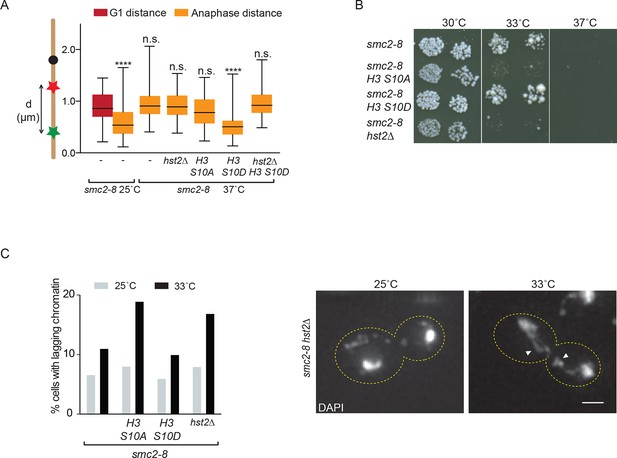
Condensin and HST2 are in the same genetic pathway that ensures proper chromosome segregation.
(A) TRP1-LYS4 distances were determined for the indicated strains in anaphase after shifting cells for 90 min to 37ºC. Box shows median value, whiskers all data points n>30. One way ANOVA was performed to test significance between G1 in smc2-8 at 25ºC and other strains, **** p<0.0001 and n.s. not significant. (B) Spotting assay of indicated strains on YPD plates at the indicated temperatures. (C) Percentage of anaphase cells containing anaphase bridges for the indicated strains after 90 min at 25ºC or 90 min at 33ºC. DAPI, 4', 6-diamidino-2-phenylindole; YPD, yeast extract peptone dextrose. n>240, scale bar is 2 µm.
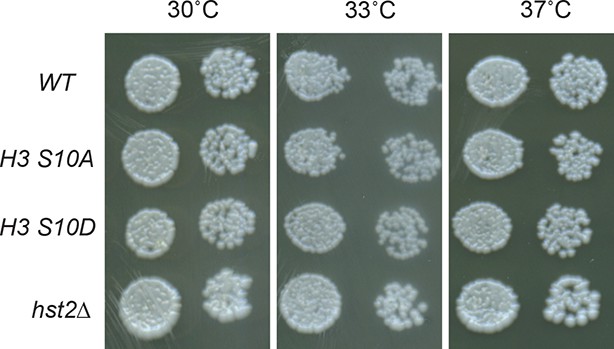
Plate growing spotting assay on YPD plates of wild type, H3 S10A, H3S10D and hst2Δ strains at different temperatures.
YPD, yeast extract peptone dextrose.
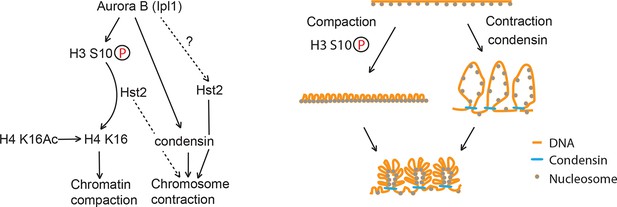
Model of chromosome condensation.
H3 S10 phosphorylation leads to chromatin fiber closure, whereas condensin is required for axial shortening. Hst2 is needed for both of these levels of condensation.
Tables
Yeast strains used in this study
| yYB number | Mating type | genotype |
|---|---|---|
| N/A | a | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 smc2::smc2-8:kanMX |
| N/A | a | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| 3476 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP |
| 3477 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP ipl1-321 |
| 4699 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP Spc42-GFP:hph hht1::hht1-S10A:KanMXloxP hht2::hht2-S10A:bleloxP ura3 ade1 leu2 |
| 8080 | a | Nup170-GFP:HIS3 ura3 ade1 leu2 |
| 9101 | a | his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 can1::MFA1pr-HIS3 hht1-hhf1::NatMX4 hht2-hhf2::[HHTS-HHFS]-URA3 |
| 9332 | a | his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 can1::MFA1pr-HIS3 hht1-hhf1::NatMX4 hht2-hhf2::[HHT-S10D-HHFS]-URA3 |
| 10818 | a | his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 can1::MFA1pr-HIS3 hht1-hhf1::NatMX4 hht2-hhf2::[HHTS10AS]-URA3 |
| 11676 | a | smc2-8 hhf1-hht1::NatMX hst2::hphNT1 ura3 ade1 leu2 |
| 12002 | a | hst2::KanMX his3∆1 leu2∆0 ura3∆0 met15∆0 |
| 10116, 10117 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP Hhf1-Hht1::NatMX hht2-hhf2::[HHT-HHF∆9-16]-URA3 ura3 ade1 leu2 |
| 10122, 10123 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP Hhf1-Hht1::NatMX hht2-hhf2::[HHT-HHFK16R]-URA3 ura3 ade1 leu2 |
| 10331 | a | Nup170-GFP:HIS3 hst2::NatMX ura3 ade1 leu2 |
| 11518, 11519 | diploid | trp1::TetO:TRP1/TRP1 lys4::LacO:LEU2/LYS4 TetR-mRFP leu2/leu2 ura3/ura3 his3/his3 |
| 11520, 11521 | diploid | trp1::TetO:TRP1/TRP1 TetR-mRFP/ TetR-GFP:LEU2 leu2/leu2 ura3/ura3 his3/his3 |
| 11173, 11989 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP hht1::hht1-S10A:KanMXloxP hht2::hht2-S10A:bleloxP smc2-8 ura3 ade leu2 |
| 9782, 9783, 9784 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP Hhf1-Hht1::NatMX ura3 ade1 leu2 |
| 10006, 10007, 10008 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP Hhf1-Hht1::NatMX hst2::hphNT1 ura3 ade1 leu2 |
| 10274, 10276, 10277 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP smc2-8 |
| 10450, 10451, 10452 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP HHT2S10D:URA3 ura3 ade1 leu2 |
| 10584, 10585, 10586 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP HHT2S10D:URA3 smc2-8 |
| 10858, 10859, 10860 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP HHT2S10D:URA3 hst2::NatMX ura3 ade1 leu2 |
| 10861, 10862, 10863 | a | trp1::TetO:TRP1lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP HHT2S10D:URA3 smc2-8 hst2::NatMX |
| 11566, 11567, 11568 | alpha | his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 can1::MFA1pr-HIS3 hht1-hhf1::NatMX4 hht2-hhf2::[HHTS10AS]-URA3 smc2-8 |
| 11767, 11768, 11769 | a, alpha | his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 can1::MFA1pr-HIS3 hht1-hhf1::NatMX4 hht2-hhf2::[HHTS-HHFS]-URA3 smc2-8 |
| 12155, 12156, 12157 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP hst2::KanMX |
| 12731, 12732, 12733 | a | his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 can1::MFA1pr-HIS3 hht1-hhf1::NatMX4 hht2-hhf2::[HHT-S10D-HHFS]-URA3 smc2-8 |
| 13100,13102 | a | trp1::TetO:TRP1 lys4::LacO:LEU2 his3::LacR-GFP:HIS3 TetR-mRFP Hhf1-Hht1::NatMX hht2-hhf2::[HHT-HHFK16R]-URA3 hst2::hphNT1 ura3 ade1 leu2 |





