Yerba mate (Ilex paraguariensis) genome provides new insights into convergent evolution of caffeine biosynthesis
Figures
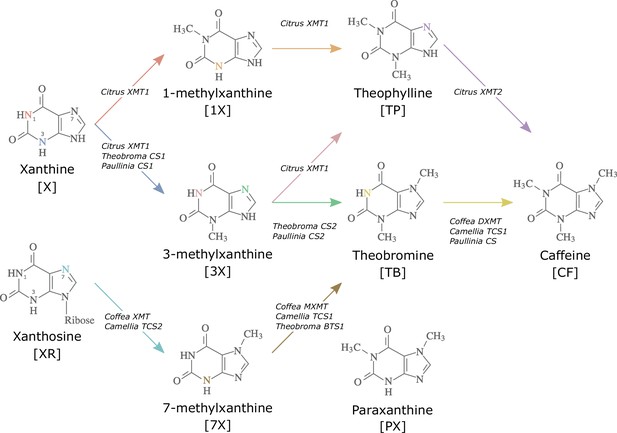
Biosynthetic routes to caffeine within the xanthine alkaloid network.
CF, caffeine; PX, paraxanthine; TB, theobromine; TP, theophylline; 1X, 1-methylxanthine; 3X, 3-methylxanthine; 7X, 7-methylxanthine; XR, xanthosine; X, xanthine. Nitrogen atoms are coloured to match the arrows corresponding to the enzymes that methylate them. Adapted with permission from O’Donnell et al., 2021.
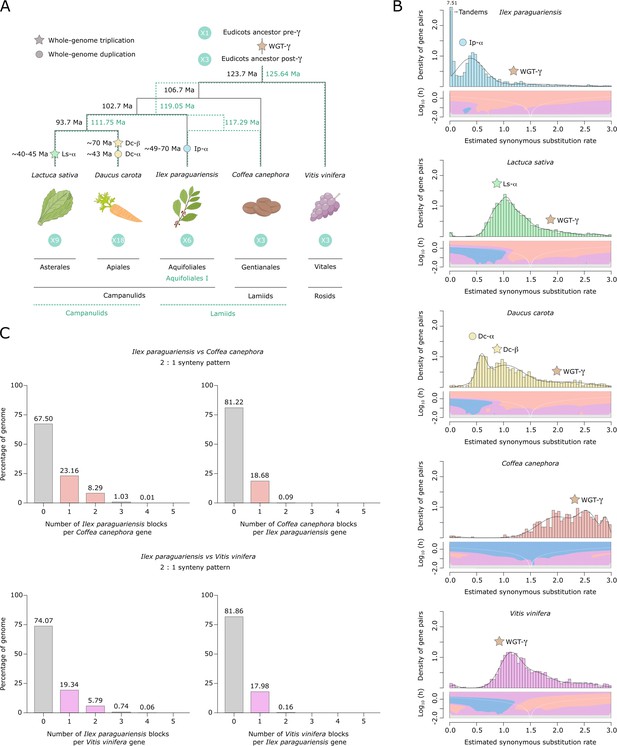
Yerba mate genome duplication history.
(A) Evolutionary scenario of the eudicot genomes of Lactuca sativa, Daucus carota, Ilex paraguariensis, Coffea canephora, and Vitis vinifera, from their ancestor pre-γ. The plastid genome phylogeny is represented with solid black lines, while the multiple nuclear genome phylogeny is represented with green dashed lines. Paleopolyploidizations are shown with coloured dots (duplications) and stars (triplications). Divergence time estimates for the lineages, as well as age estimates for the L. sativa and D. carota paleopolyploidizations were obtained from the literature (Iorizzo et al., 2016; Magallón et al., 2015; Reyes-Chin-Wo et al., 2017; Zhang et al., 2020b). Ma, million years ago. (B) Ks distributions with Gaussian mixture model and SiZer analyses of I. paraguariensis (blue), L. sativa (green), D. carota (yellow), C. canephora (red), and V. vinifera (purple) paralogues. SiZer maps below histograms identify significant peaks at corresponding Ks values. Blue represents significant increases in slope, red indicates significant decreases, purple represents no significant slope change, and grey indicates not enough data for the test. (C) Comparative genomic synteny analyses of I. paraguariensis with C. canephora and V. vinifera.
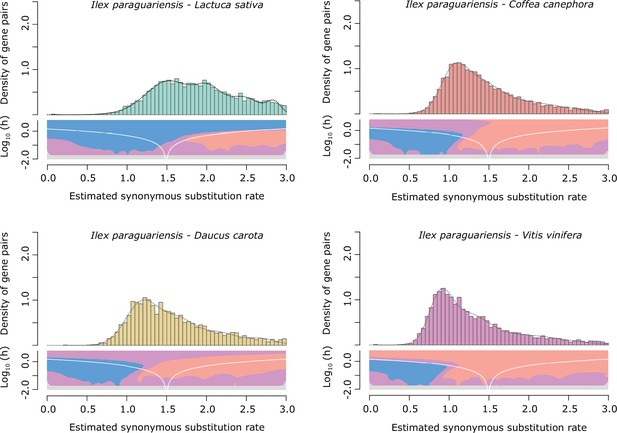
Ks distributions with Gaussian mixture model and SiZer analyses of I. paraguariensis and L. sativa (green), D. carota (yellow), C. canephora (red), and V. vinifera (purple) orthologues.
SiZer maps below histograms identify significant peaks at corresponding Ks values. Blue represents significant increases in slope, red indicates significant decreases, purple represents no significant slope change, and grey indicates not enough data for the test.
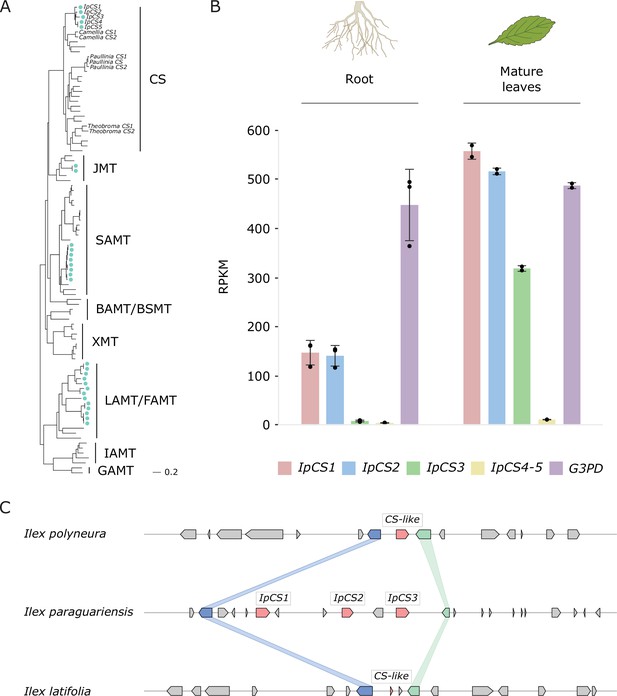
The yerba mate (YM) genome encodes three recently duplicated CS-type SABATH proteins that are expressed in caffeine-producing tissues.
(A) SABATH gene tree estimate (LnL = −34,265.473) shows the placement of full-length YM proteins (marked by blue-green dots) within clades that have published functions. GAMT, gibberellin MT; IAMT, indole-3-acetic acid MT; LAMT/FAMT, loganic/farnesoic acid MT; BAMT/BSMT, benzoic/salicylic acid MT; XMT, xanthine alkaloid MT used for caffeine biosynthesis in Coffea and Citrus; SAMT, salicylic acid MT; JMT, jasmonic acid MT; CS, caffeine synthase in Theobroma, Camellia, and Paullinia. Accession numbers for all sequences are provided in Figure 3—source data 1. (B) Gene expression analysis of IpCS1–5 in root (n = 3) and mature leaves (n = 2) as indicated by the relative abundance of YM transcriptome reads mapped to the IpCS1–5 transcripts. RPKM, reads per kilobase per million mapped reads. Error bars indicate standard deviation from the mean. Housekeeping gene: G3PD, glyceraldehyde-3-phosphate dehydrogenase. (C) Synteny-based analysis of the CS genomic region for I. paraguariensis, I. polyneura, and I. latifolia.
-
Figure 3—source data 1
Accession numbers of SABATH sequences used for phylogenetic analysis in Figure 3.
- https://cdn.elifesciences.org/articles/104759/elife-104759-fig3-data1-v1.docx
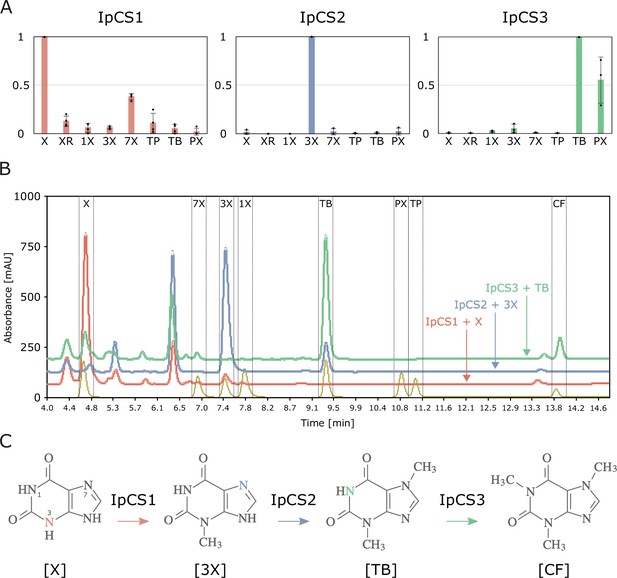
SABATH enzymes have evolved to catalyse the biosynthesis of caffeine in yerba mate.
(A) Relative enzyme activitiy of IpCS1 (n = 4), IpCS2 (n = 3), and IpCS3 (n = 3) SABATH enzymes with eight xanthine alkaloid substrates. (B) High-performance liquid chromatography (HPLC) traces showing products formed by three encoded caffeine synthase (CS)-type enzymes. Absorbance at 254 nm is shown. (C) Proposed biosynthetic pathway for caffeine in yerba mate. X, xanthine; XR, xanthosine; 1X, 1-methylxanthine; 3X, 3-methylxanthine; 7X, 7-methylxanthine; TP, theophylline; TB, theobromine; PX, paraxanthine. Coloured atoms and arrows indicate atoms that act as methyl acceptors for a given reaction.
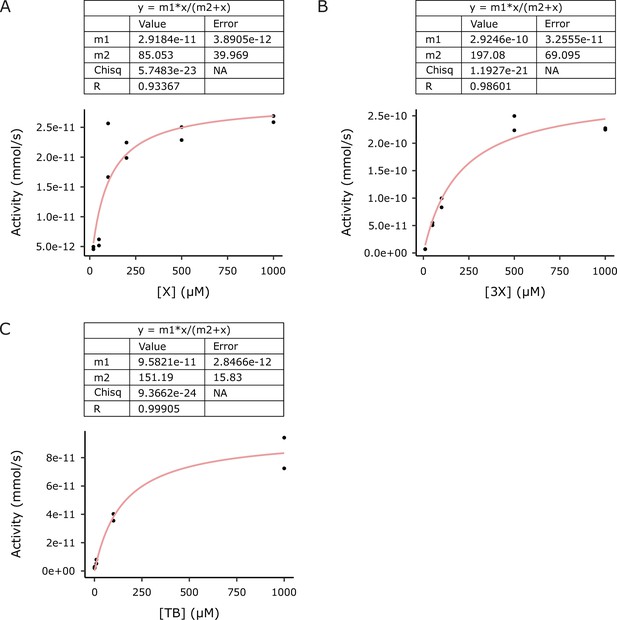
Michaelis–Menten curves used to estimate kinetic parameters for (A) IpCS1, (B) IpCS2, and (C) IpCS3 (n = 2).
m1 is the estimate for Vmax and m2 is the estimate for KM. X, xanthine; 3X, 3-methylxanthine; TB, theobromine.
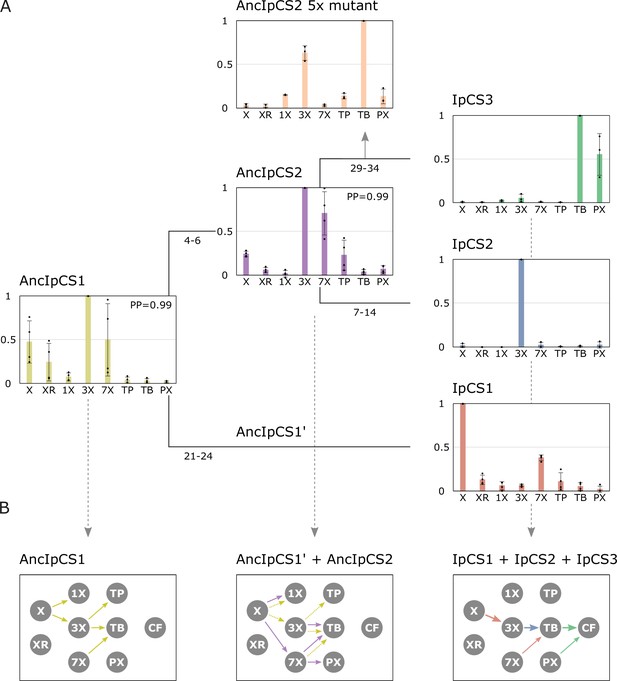
Ancestral sequence resurrection reveals ancestral xanthine alkaloid pathway flux.
(A) Simplified evolutionary history of three yerba mate (YM) xanthine alkaloid-methylating enzymes and their two ancestors, AncIpCS1 and AncIPCS2. Average site-specific posterior probabilities (PP) for each ancestral enzyme estimate are provided. Numbers below each branch of the phylogeny represents the number of amino acid replacements between each enzyme shown. These two ancestral relative activity charts (n = 4) show the averaged activities of two allelic variants of each enzyme. Relative substrate preference is also shown for the AncIPCS2 mutant enzyme (n = 3) in which five amino acid positions, A22G, R23C, T25S, H221N, and Y265C, that are inferred to have been replaced during the evolution of IpCS3, were changed. (B) Inferred pathway flux is shown for the antecedent pathways that could have been catalysed by the ancestral or modern-day combinations of enzymes that would have existed at three time points in the history of the enzyme lineage. Arrows linking metabolites are coloured according to the activities detected from each enzyme shown in panel A. Dotted arrows are shown for AncIpCS1’ because it is unknown what characteristics it would possess; it is assumed that it would have at least catalysed the formation of 3X from X since both its ancestor and descendant enzyme do so. X, xanthine; XR, xanthosine; 1X, 1-methylxanthine; 3X, 3-methylxanthine; 7X, 7-methylxanthine; TP, theophylline; TB, theobromine; PX, paraxanthine.
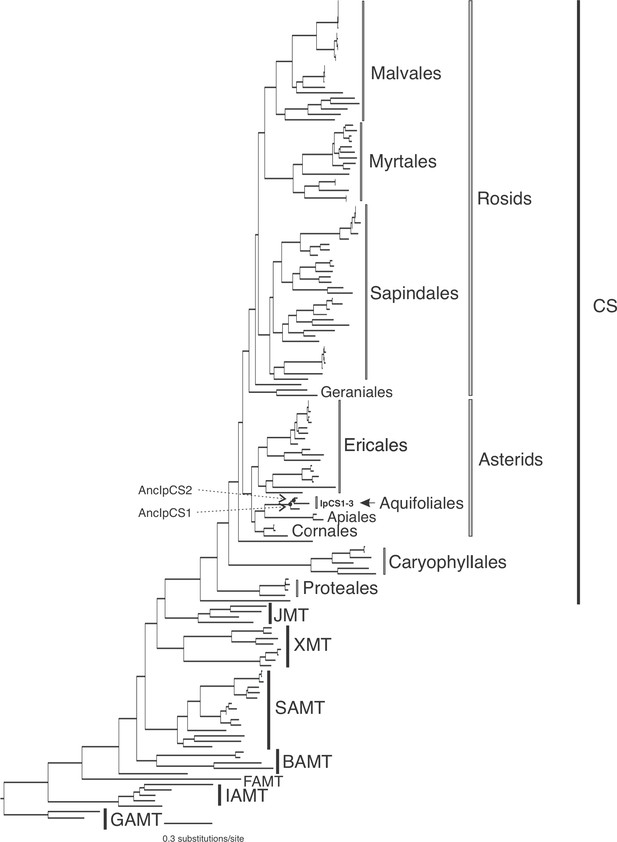
SABATH enzyme family phylogenetic tree used for obtaining ancestral sequence estimates for the clade including IpCS1–3 of Aquifoliales (log-likelihood = −46,631.672).
Clades of enzymes for which at least one sequence has been functionally characterized are labelled. GAMT, gibberellin MT; IAMT, indole-3-acetic acid MT; FAMT, farnesoic acid MT; BAMT, benzoic acid MT; XMT, xanthine alkaloid MT used for caffeine biosynthesis in Coffea and Citrus; SAMT, salicylic acid MT; JMT, jasmonic acid MT; CS, caffeine synthase in Theobroma, Camellia, and Paullinia. Within the CS clade, the orders of rosids and asterids are labelled to show interrelationships.
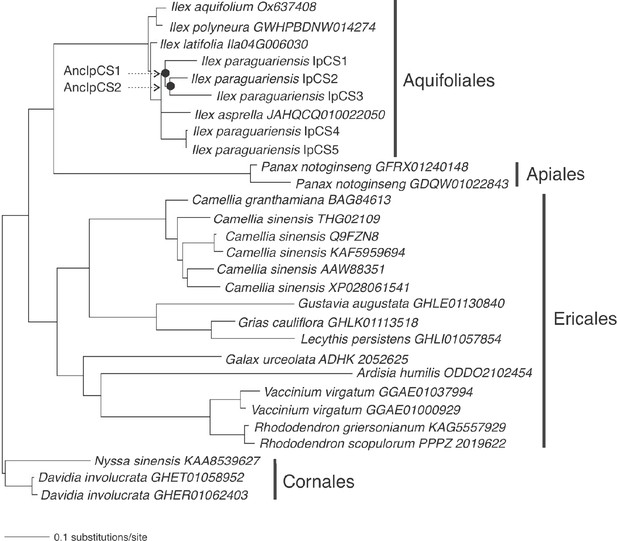
Caffeine synthase enzyme family phylogenetic tree used for obtaining alternative ancestral sequence estimates for AncIpCS1 and 2 (log-likelihood = −7032.8928).

Alignment of the two estimated amino acid sequences for AncIpCS1 that were biochemically characterized in Figure 5.

Alignment of the two estimated amino acid sequences for AncIpCS2 that were biochemically characterized in Figure 5.
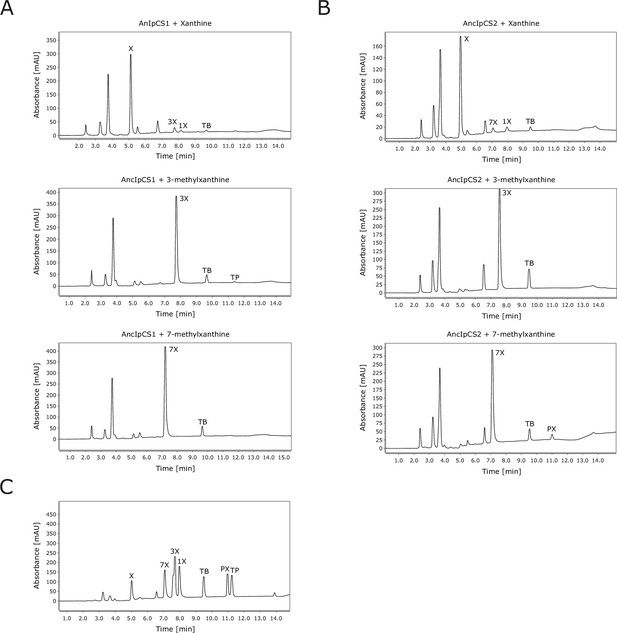
High-performance liquid chromatography (HPLC) traces for xanthine alkaloid products formed by ancestral Ilex caffeine synthase (CS) enzymes.
(A) Chromatograms for AncIpCS1 assayed with three substrates. (B) Chromatograms for AncIpCS2 assayed with three substrates. (C) Chromatogram for authentic standards. X, xanthine; XR, xanthosine; 1X, 1-methylxanthine; 3X, 3-methylxanthine; 7X, 7-methylxanthine; TP, theophylline, TB, theobromine; PX, paraxanthine. Absorbance at 254 nm is shown.
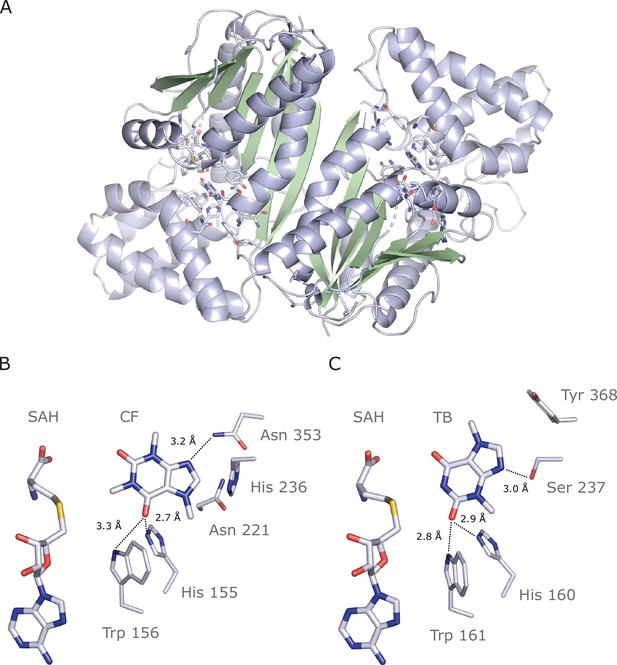
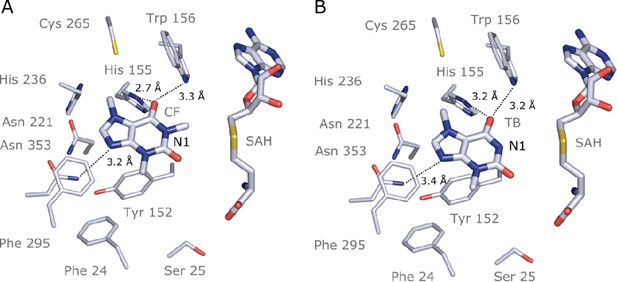
Crystal structure of IpCS3 in complex with caffeine (CF) and S-adenosyl-homocysteine (SAH) and comparison with the active site of Coffea canephora DXMT.
(A) Overview of the crystal structure of IpCS3 (PDB ID: 8UZD) depicting the active site of the enzyme in complex with CF and SAH. (B) Relevant residues in IpCS3 for ligand recognition. (C) Relevant residues in CcDXMT (PDB ID: 2EFJ) for ligand recognition. Protein residues are displayed as lines with carbon atoms coloured in bluewhite while small molecules – CF, theobromine (TB), and SAH – are drawn as sticks. Colour code for the rest of the atoms: nitrogen (blue), oxygen (red), and sulphur (yellow). Hydrogen bond interactions are indicated as black dotted lines.
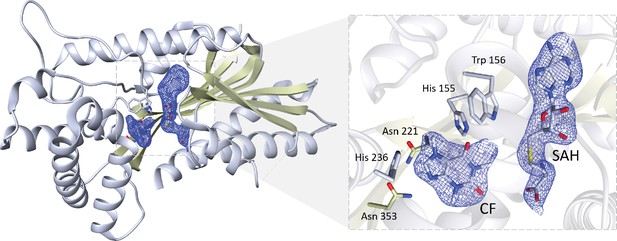
Crystal structure of IpCS3 displaying a difference Fourier map (Fo − Fc) contoured to 2.0 σ (blue) showing bound SAH and CF.
Relevant residues in IpCS3 for ligand recognition are displayed as lines with carbon atoms coloured in grey, while small molecules – caffeine (CF) and S-adenosyl-homocysteine (SAH) – are drawn as sticks and labelled. Colour code for the rest of the atoms: nitrogen (blue), oxygen (red), and sulphur (yellow).
Theobromine and caffeine are oriented the same way in the active site of IpCS3.
(A) IpCS3–CF complex (PDB ID: 8T2G). (B) IpCS3–TB complex (docking model). Protein residues are displayed as lines with carbon atoms coloured in bluewhite while small molecules – theobromine (TB), caffeine (CF), and S-adenosyl-homocysteine (SAH) – are drawn as sticks. Colour code for the rest of the atoms: nitrogen (blue), oxygen (red), and sulphur (yellow). Hydrogen bond interactions are indicated as dotted lines.
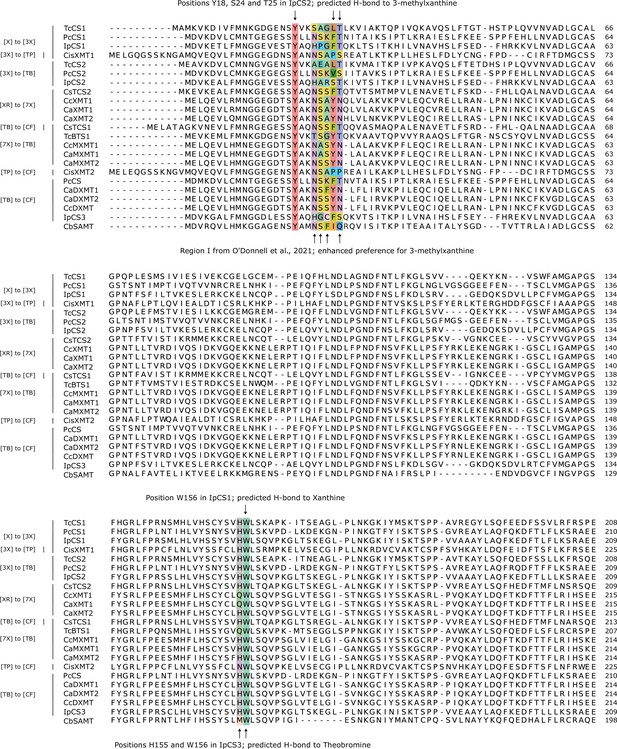
Comparative amino acid alignment of xanthine methyltransferase (XMT) and caffeine synthase (CS) sequences (1–209 of IpCS1) shows convergent changes predicted to participate in substrate binding and promote methylation preference switches.
Accession numbers are as follows: TcCS1 (EOY17874), PcCS1 (EC766748), IpCS1 (CAK9135737), CisXMT1 (KDO50937), TcCS2 (EOY17880), PcCS2 (EC778019), IpCS2 (CAK9135740), CsTCS2 (AB031281), CcXMT1 (JX978518), CaXMT1 (AB048793), CaXMT2 (AB084127), CsTCS1 (AB031280), TcBTS1 (AB096699), CcMXMT1 (JX978517), CaMXMT1 (AB048794), CaMXMT2 (AB084126), CisXMT2 (XP_006469448), PcCS (BK008796), CaDXMT1 (AB084125), CaDXMT2 (KJ577793), CcDXMT (JX978516), IpCS3 (CAK9135742), and CbSAMT (AAF00108).
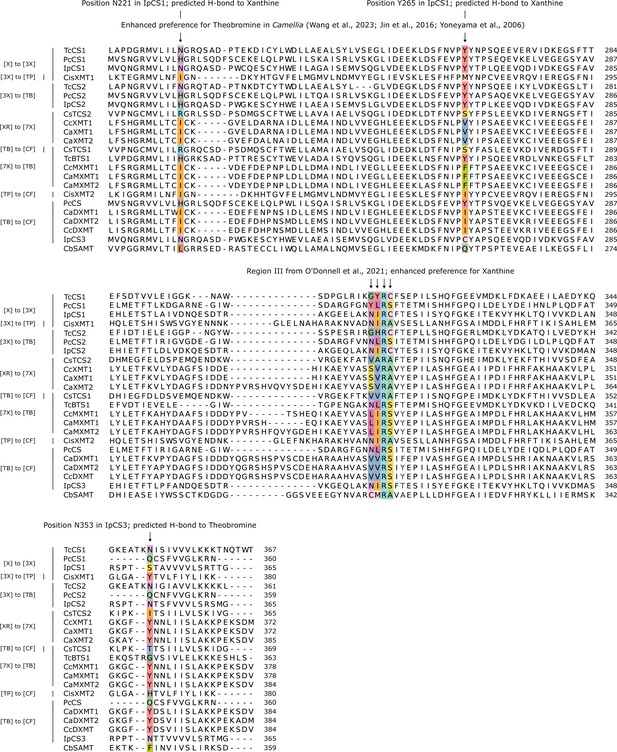
Comparative amino acid alignment of xanthine methyltransferase (XMT) and caffeine synthase (CS) sequences (210–365 of IpCS1) shows convergent changes predicted to participate in substrate binding and promote methylation preference switches.
Accession numbers are as follows: TcCS1 (EOY17874), PcCS1 (EC766748), IpCS1 (CAK9135737), CisXMT1 (KDO50937), TcCS2 (EOY17880), PcCS2 (EC778019), IpCS2 (CAK9135740), CsTCS2 (AB031281), CcXMT1 (JX978518), CaXMT1 (AB048793), CaXMT2 (AB084127), CsTCS1 (AB031280), TcBTS1 (AB096699), CcMXMT1 (JX978517), CaMXMT1 (AB048794), CaMXMT2 (AB084126), CisXMT2 (XP_006469448), PcCS (BK008796), CaDXMT1 (AB084125), CaDXMT2 (KJ577793), CcDXMT (JX978516), IpCS3 (CAK9135742), and CbSAMT (AAF00108).
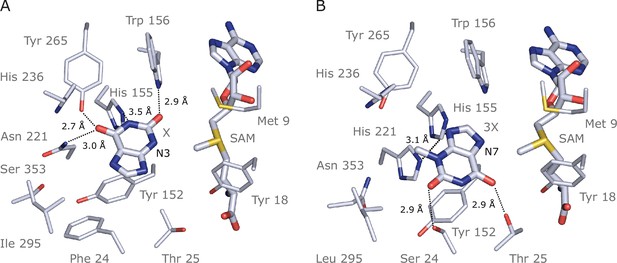
Docking models of xanthine alkaloids in IpCS1 and IpCS2 active sites.
(A) IpCS1–X complex. (B) IpCS2–3X complex. Protein residues are displayed as lines with carbon atoms coloured in bluewhite while small molecules – xanthine (X), 3-methylxanthine (3X), caffeine (CF), paraxanthine (PX), S-adenosyl-L-methionine (SAM), and S-adenosyl-homocysteine (SAH) – are drawn as sticks. Colour code for the rest of the atoms: nitrogen (blue), oxygen (red), and sulphur (yellow). Hydrogen bond interactions are indicated as black dotted lines.
AlphaFold2-ColabFold Model Quality assessment of IpCS1, IpCS2, and IpCS3 models.
Sequence coverage of the multiple sequence alignment used for IpCS1 (A), IpCS2 (B), and IpCS3 (C). Alignment error for IpCS1 (D), IpCS2 (E), and IpCS3 (F). pLDDT score of IpCS1 (G), IpCS2 (H), and IpCS3 (I).
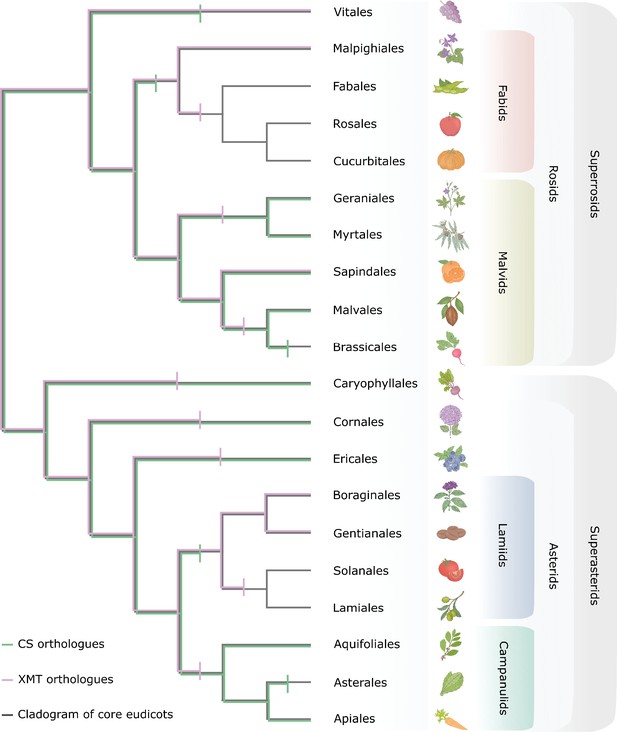
Only CS genes were available for co-option and utilization for xanthine alkaloid biosynthesis in yerba mate whereas coffee only had xanthine methyltransferase (XMT) genes.
Both CS- and XMT-type caffeine biosynthetic enzymes were present in the ancestor of core eudicots but numerous apparent losses of one or the other or both has occurred during lineage diversification. Gene loss is represented by vertical bar on relevant branches of the cladogram.
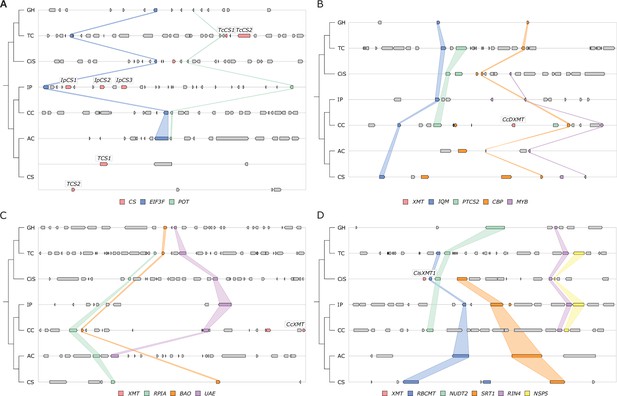
Only CS genes are available for co-option and utilization for xanthine alkaloid biosynthesis in yerba mate.
(A) Synteny-based analysis of the CS genomic region for seven angiosperm taxa. (B–D) Synteny-based analyses of the XMT genomic regions for seven angiosperm taxa. Angiosperm taxa: GH, Gossypium hirsutum; TC, Theobroma cacao; CiS, Citrus sinensis; IP, Ilex paraguariensis; CC, Coffea canephora; AC, Actinidia chinensis; CS, Camellia sinensis. Genes: CS, caffeine synthase-type enzyme; EIF3F, eukaryotic translation initiation factor 3 subunit F; POT, proton‐dependent oligopeptide transporter; XMT, xanthine methyltransferase-type enzyme; RPIA, ribose 5-phosphate isomerase A; BAO, beta-amyrin 28-oxidase-like; UAE, UDP-arabinose 4-epimerase 1-like; IQM, IQ domain-containing protein; PTC52, protochlorophyllide-dependent translocon component 52; CBP, calcium-binding protein; MYB, MYB transcription factor; RBCMT, ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase; NUDT2, nudix hydrolase 2-like, SRT1, NAD-dependent protein deacetylase; RIN4, RPM1 interacting protein 4; NSP5, nitrile specifier protein 5.
Tables
Statistics of the genome sequencing data of yerba mate.
| Library | Number of reads | Read length | Total length | Coverage |
|---|---|---|---|---|
| Pair-end 350 bp #1 | 360,653,408 | 101 | 36.4 Gbp | 21.8× |
| Pair-end 350 bp #2 | 368,746,464 | 101 | 37.2 Gbp | 22.3× |
| Pair-end 550 bp | 356,261,246 | 101 | 36 Gbp | 21.5× |
| Mate-pair 3 kbp #1 | 415,398,586 | 101 | 30.3 Gbp | 18.2× |
| Mate-pair 3 kbp #2 | 410,588,934 | 101 | 30 Gbp | 17.9× |
| Mate-pair 3 kbp #3 | 343,059,350 | 101 | 25 Gbp | 15× |
| Mate-pair 8 kbp | 393,202,256 | 101 | 34.6 Gbp | 20.7× |
| Mate-pair 12 kbp | 415,478,776 | 101 | 33.7 Gbp | 20.1× |
| PacBio long reads | 19,514,627 | 50 bp to 61 kbp | 77.5 Gbp | 49.3× |
| Total | 341 Gbp | 207.8× |
Statistics of the genome assembly of yerba mate.
| Metric | Value |
|---|---|
| # scaffolds (≥1000 bp) | 10,611 |
| # scaffolds (≥5000 bp) | 9343 |
| # scaffolds (≥10,000 bp) | 8951 |
| # scaffolds (≥25,000 bp) | 5944 |
| # scaffolds (≥50,000 bp) | 2595 |
| Total length (≥50,000 bp) | 887,124,725 |
| # scaffolds | 10,611 |
| Largest scaffold | 7,402,063 |
| Total length | 1,064,802,823 |
| GC (%) | 36.33 |
| N50 | 510,878 |
| N75 | 132,523 |
| L50 | 506 |
| L75 | 1461 |
| # N’s per 100 kbp | 1976.99 |
Classification and distribution of repetitive DNA elements in yerba mate.
| Number | Length occupied (bp) | Percentage of the genome (%) | |
|---|---|---|---|
| Class I retrotransposons | 421,599 | 385,714,532 | 36.22 |
| SINEs | 840 | 154,298 | 0.01 |
| Penelope | 0 | 0 | 0.00 |
| LINEs | 35,433 | 17,109,207 | 1.61 |
| CRE/SLACS | 0 | 0 | 0.00 |
| L2/CR1/Rex | 575 | 135,549 | 0.01 |
| R1/LOA/Jockey | 443 | 76,937 | 0.01 |
| R2/R4/NeSL | 0 | 0 | 0.00 |
| RTE/Bov-B | 8599 | 2,126,765 | 0.20 |
| L1/CIN4 | 25,816 | 14,769,956 | 1.39 |
| LTR retrotransposons | 385,326 | 368,451,027 | 34.60 |
| BEL/Pao | 709 | 266,632 | 0.03 |
| Ty1/Copia | 98,237 | 67,631,136 | 6.35 |
| Gypsy/DIRS1 | 216,472 | 274,526,515 | 25.78 |
| Retroviral | 0 | 0 | 0.00 |
| Class II DNA transposons | 45,427 | 19,116,209 | 1.80 |
| hobo-Activator | 21,335 | 6,378,850 | 0.60 |
| Tc1-IS630-Pogo | 0 | 0 | 0.00 |
| En-Spm | 0 | 0 | 0.00 |
| MuDR-IS905 | 0 | 0 | 0.00 |
| PiggyBac | 0 | 0 | 0.00 |
| Tourist/Harbinger | 5870 | 2,846,548 | 0.27 |
| Others | 0 | 0 | 0.00 |
| Unclassified | 990,080 | 269,430,122 | 25.30 |
| Total interspersed repeats | 674,260 | 863 | 63.32 |
| Small RNA | 4362 | 718,762 | 0.07 |
| Satellites | 0 | 0 | 0.00 |
| Simple repeats | 185,507 | 7,911,080 | 0.74 |
| Low complexity | 31,856 | 1,606,255 | 0.15 |
Apparent enzyme kinetic parameter estimates for yerba mate caffeine biosynthetic enzymes with selected substrates.
| Enzyme (substrate) | KM (μM) | kcat (1/s) | kcat/KM (s–1 M–1) |
|---|---|---|---|
| IpCS1 (X) | 85.05 | 0.0009 | 10.11 |
| IpCS2 (3X) | 197.08 | 0.0031 | 15.77 |
| IpCS3 (TB) | 151.19 | 0.0029 | 19.36 |
Data collection and refinement statistics of IpCS3 structure bound to S-adenosyl-homocysteine (SAH) and caffeine.
| IpCS3 in complex with SAH and caffeine | |
|---|---|
| PDB | 8UZD |
| Data collection | |
| Wavelength (Å) | 0.9786 |
| Resolution (Å) | 2.72 |
| Resolution rangea* | 37.00–2.72 |
| (2.82–2.72) | |
| Space group | P 41 21 2 |
| Cell dimensions | |
| a, b, c (Å) | 82.67, 82.67, 226.09 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Total reflections | 43,818 |
| Unique reflections | 21,910 |
| Multiplicitya* | 2.0 (2.0) |
| Completeness (%)a* | 99.89 (100.00) |
| <I/σI>a | 25.79 (2.87) |
| Rmergea,b†* (%) | 0.0223 (0.2168) |
| Rmeas (%)a* | 0.0315 (0.3066) |
| CC1/2a* | 0.999 (0.878) |
| Refinement | |
| Resolution (Å) | 2.72 |
| No. reflections | 21,909 |
| Rworkc ‡/Rfreed § | 0.194/0.248 |
| No. atoms | |
| Protein | 5,216 |
| CFF + SAH | 80 |
| Water | 48 |
| B-factors | |
| Protein | 63.38 |
| CFF + SAH | 84.48 |
| Water | 48.19 |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 1.112 |
-
*
aNumbers in parentheses refer to the highest resolution shell.
-
†
bRmerge = Σ|Ii − <Ii>|/ΣIi, where Ii = the intensity of the ith reflection and <Ii> = mean intensity.
-
‡
cRwork = Σ|Fo − Fc|/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
-
§
dRfree was calculated as for Rwork, but on a test set comprising 5% of the data excluded from refinement.
Detail of yerba mate tRNA and anti-codon nucleotide sequences.
| tRNA genes | Anti-codon counts | Total No. of tRNAs | |||||
|---|---|---|---|---|---|---|---|
| POLAR | |||||||
| Asparagine (Asn) | GTT (36) | ATT (0) | 36 | ||||
| Cysteine (Cys) | GCA (22) | ACA (0) | 22 | ||||
| Glutamine (Gln) | TTG (13) | CTG (10) | 23 | ||||
| Glycine (Gly) | GCC (32) | TCC (11) | CCC (8) | ACC (0) | 51 | ||
| Serine (Ser) | GCT (20) | TGA (20) | AGA (15) | CGA (5) | GGA (5) | ACT (0) | 65 |
| Threonine (Thr) | TGT (11) | AGT (16) | GGT (6) | CGT (2) | 35 | ||
| Tyrosine (Tyr) | GTA (17) | ATA (0) | 17 | ||||
| NON-POLAR | |||||||
| Alanine (Ala) | AGC (12) | CGC (4) | TGC (11) | GGC (0) | 27 | ||
| Isoleucine (Ile) | AAT (14) | TAT (6) | GAT (2) | 22 | |||
| Leucine (Leu) | CAA (23) | AAG (10) | CAG (4) | TAG (8) | TAA (6) | GAG (0) | 51 |
| Methionine (Met) | CAT (55) | 55 | |||||
| Phenylalanine (Phe) | GAA (30) | AAA (2) | 32 | ||||
| Proline (Pro) | AGG (10) | TGG (28) | CGG (4) | GGG (0) | 42 | ||
| Tryptophan (Trp) | CCA (31) | 31 | |||||
| Valine (Val) | AAC (11) | GAC (10) | CAC (9) | TAC (7) | 37 | ||
| POSITIVELY CHARGED | |||||||
| Arginine (Arg) | ACG (15) | TCT (14) | CCT (7) | CCG (6) | TCG (6) | GCG (3) | 51 |
| Histidine (His) | GTG (25) | ATG (2) | 27 | ||||
| Lysine (Lys) | CTT (10) | TTT (17) | 27 | ||||
| NEGATIVELY CHARGED | |||||||
| Aspartic acid (Asp) | GTC (39) | ATC (1) | 40 | ||||
| Glutamic acid (Glu) | CTC (14) | TTC (21) | 35 | ||||
| Selenocysteine tRNAs | TCA (0) | 0 | |||||
| Possible suppressor tRNAs | CTA (0) | TTA (1) | TCA (1) | 2 | |||
| tRNAs with undetermined isotypes | 11 | ||||||
| Predicted pseudogenes | 76 | ||||||
miRNA families predicted in the yerba mate genome.
| miRNA | Functional involvement in other eudicot plants |
|---|---|
| miR156 | Seed growth and development Chi et al., 2011; Song et al., 2011 |
| Fruit development (Pantaleo et al., 2010) | |
| Drought/cold stress (Curaba et al., 2012; Zhu and Luo, 2013) | |
| miR159 | Growth and development (Varkonyi-Gasic et al., 2010) |
| Phase change from vegetative to reproductive growth (Han et al., 2014) | |
| Lipid and protein accumulation (Zhao et al., 2010) | |
| Drought stress (Barrera-Figueroa et al., 2011) | |
| miR160 | Growth and development (Gu et al., 2013; Wang et al., 2011) |
| Fibrous root and storage root development (Sun et al., 2015) | |
| Drought stress (Nadarajah and Kumar, 2019) | |
| miR162_2 | Storage root initiation and development (Sun et al., 2015) |
| miR164 | Lateral root and leaf development (Deng et al., 2015) |
| Fibrous root and storage root development (Sun et al., 2015) | |
| Seed development (Song et al., 2011) | |
| Drought stress (Ferreira et al., 2012) | |
| miR166 | Seed development (Song et al., 2011) |
| Fibrous root and storage root development (Sun et al., 2015) | |
| Drought stress (Barrera-Figueroa et al., 2011) | |
| Disease resistance (Guo et al., 2011) | |
| miR167_1 | Growth and development (Varkonyi-Gasic et al., 2010) |
| Drought/cold stress (Barrera-Figueroa et al., 2011; Jeong et al., 2011) | |
| miR168 | Development (Gu et al., 2013) |
| Resistance to fire blight (Kaja et al., 2015) | |
| miR169_2; miR169_5 | Drought/cold/salt stress (Carnavale Bottino et al., 2013; Koc et al., 2015; Sheng et al., 2015; Shui et al., 2013) |
| miR171_1; miR171_2 | Development (Chaves et al., 2015; Zhang et al., 2011) |
| Lipid and protein accumulation (Zhao et al., 2010) | |
| miR172 | Development (Sun et al., 2012) |
| Starch biosynthesis (Chen et al., 2015) | |
| Drought/cold stress (Koc et al., 2015) | |
| miR390 | Drought stress (Shui et al., 2013) |
| Leaf morphology (Karlova et al., 2013) | |
| miR394 | Drought/salt stress (Song et al., 2013) |
| miR395 | Low sulfate response (Katiyar et al., 2012) |
| miR396 | Seed development (Gao et al., 2015) |
| Starch biosynthesis (Chen et al., 2015) | |
| Drought/salt stress (Shui et al., 2013; Xie et al., 2014) | |
| miR397 | Drought/cold stress (Koc et al., 2015) |
| miR398 | Fibrous root and storage root development (Sun et al., 2015) |
| Salt stress (Carnavale Bottino et al., 2013) | |
| miR399 | Phosphate homeostasis (Katiyar et al., 2012; Pant et al., 2008) |
| Shoot to root transport (Pant et al., 2008) | |
| miR403 | Drought stress (Shui et al., 2013) |
| miR405 | Transposon derived (Xie et al., 2005) |
| miR408 | Tolerance to Boron deficiency (Lu et al., 2015) |
| Cold stress (Zhang et al., 2014) | |
| Response to wounding and topping (Tang et al., 2012) | |
| miR473 | Metabolism (Din et al., 2014) |
| Stress response (Patanun et al., 2013) | |
| miR474 | Drought stress (Kantar et al., 2011) |
| miR475 | Metabolism (Din et al., 2014) |
| miR477 | Starch biosynthesis (Xie et al., 2011) |
| miR530 | Disease resistance (Zhao et al., 2015) |
| miR1023 | Disease resistance (Jiao and Peng, 2018) |
| miR1446 | Stress response (Lu et al., 2008) |
miRNA targets predicted in the yerba mate genome.
| Targets IDs | Description | miR159 | miR164 | miR167_1 | miR168 | miR169_2 | miR169_5 | miR171_1 | miR171_2 | miR390 | miR394 | miR396 | miR397 | miR398 | miR403 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ILEXPARA_008283 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_029002 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_031381 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_043376 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_013180 | ileS, isoleucine tRNA ligase | ✸ | |||||||||||||
| ILEXPARA_000910 | myb-like transcription factor | ✸ | ✸ | ||||||||||||
| ILEXPARA_028644 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_005969 | Putative membrane protein | ✸ | |||||||||||||
| ILEXPARA_048009 | panC, pantothenate (vitamin B5) synthetase | ✸ | |||||||||||||
| ILEXPARA_018064 | arf, auxin response factor | ✸ | |||||||||||||
| ILEXPARA_019275 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_024153 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_035190 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_016483 | Hypothetical protein | ✸ | |||||||||||||
| ILEXPARA_029421 | GCP4, gamma tubulin complex protein 4 | ✸ | ✸ | ||||||||||||
| ILEXPARA_047849 | GOLS1, galactinol synthase 1 | ✸ | ✸ | ||||||||||||
| ILEXPARA_003987 | NACK1, kinesin-like protein | ✸ | ✸ | ||||||||||||
| ILEXPARA_044341 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_005359 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_035716 | RABE1C, ras-related protein | ✸ | |||||||||||||
| ILEXPARA_010316 | MAPK, mitogen activated protein kinase | ✸ | ✸ | ||||||||||||
| ILEXPARA_032923 | Uncharacterized protein | ✸ | ✸ | ||||||||||||
| ILEXPARA_008149 | Uncharacterized protein | ✸ | ✸ | ||||||||||||
| ILEXPARA_048631 | Protein kinase | ✸ | ✸ | ||||||||||||
| ILEXPARA_008152 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_023090 | NAGK, N-acetyl-D-glucosamine kinase | ✸ | |||||||||||||
| ILEXPARA_024088 | RNA-binding (RRM/RBD/RNP motif) family protein | ✸ | |||||||||||||
| ILEXPARA_023716 | Endoglucanase | ✸ | |||||||||||||
| ILEXPARA_042182 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_021515 | Pentatricopeptide repeat (PPR) protein | ✸ | |||||||||||||
| ILEXPARA_004925 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_045111 | Rotamase FKBP 1 | ✸ | |||||||||||||
| ILEXPARA_013832 | ABCC2, ABC transporter C family member 2 protein | ✸ | |||||||||||||
| ILEXPARA_039828 | guaA, GMP synthase | ✸ | |||||||||||||
| ILEXPARA_028274 | Hypothetical protein | ✸ | |||||||||||||
| ILEXPARA_024538 | RPT6A, regulatory particle triple-A ATPase 6A | ✸ | |||||||||||||
| ILEXPARA_031387 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_043757 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_005297 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_012032 | Uncharacterized protein | ✸ | |||||||||||||
| ILEXPARA_9682 | OST1B, oligosaccharyltransferase 1B | ✸ |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Ilex paraguariensis) | IpCS1 | GenBank | CAK9135737 | Xanthine methyltransferase gene of Ilex paraguariensis |
| Gene (Ilex paraguariensis) | IpCS2 | GenBank | CAK9135740 | 3-Methylxanthine methyltransferase gene of Ilex paraguariensis |
| Gene (Ilex paraguariensis) | IpCS3 | GenBank | CAK9135742 | Theobromine methyltransferase gene of Ilex paraguariensis |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Novagen | 69450-M | Chemically competent cells |
| Biological sample (Ilex paraguariensis) | Ilex paraguariensis A. St.-Hil. var. paraguariensis | INTA-EEA Cerro Azul, Misiones, Argentina | cv CA 8/74 | Used to extract genomic DNA |
| Biological sample (Ilex paraguariensis) | Ilex paraguariensis A. St.-Hil. var. paraguariensis | Establecimiento Las Marías S.A.C.I.F.A., Corrientes, Argentina | cv SI-49 | Used to extract genomic DNA |
| Recombinant DNA reagent | pUC57-IpCS1 (plasmid) | GenScript | Used to clone IpCS1 gene | |
| Recombinant DNA reagent | pTrcHis-IpCS2 (plasmid) | This paper | Used to clone IpCS2 gene | |
| Recombinant DNA reagent | pUC57-IpCS3 (plasmid) | GenScript | Used to clone IpCS3 gene | |
| Recombinant DNA reagent | pUC57-AncIpCS1 (plasmid) | GenScript | Used to clone AncIpCS1 gene | |
| Recombinant DNA reagent | pUC57-AncIpCS2 (plasmid) | GenScript | Used to clone AncIpCS2 gene | |
| Sequence-based reagent | pET-15b- IpCS1 (plasmid) | This paper | Used to express IpCS1 in E. coli BL21(DE3) | |
| Sequence-based reagent | pET-15b- IpCS2 (plasmid) | This paper | Used to express IpCS2 in E. coli BL21(DE3) | |
| Sequence-based reagent | pET-15b- IpCS3 (plasmid) | This paper | Used to express IpCS3 in E. coli BL21(DE3) | |
| Sequence-based reagent | pET-15b- AncIpCS1 (plasmid) | This paper | Used to express AncIpCS1 in E. coli BL21(DE3) | |
| Sequence-based reagent | pET-15b- AncIpCS2 (plasmid) | This paper | Used to express AncIpCS2 in E. coli BL21(DE3) | |
| Sequence-based reagent | IpCS2F | This paper | PCR primers | 5′-ATGGACGTGAAGGAAGCAC-3′ |
| Sequence-based reagent | IpCS2R | This paper | PCR primers | 5′-CTATCCCATGGTCCTGCTAAG-3′ |
| Peptide, recombinant protein | IpCS1 | This paper | Purified from E. coli BL21(DE3) cells | |
| Peptide, recombinant protein | IpCS2 | This paper | Purified from E. coli BL21(DE3) cells | |
| Peptide, recombinant protein | IpCS3 | This paper | Purified from E. coli BL21(DE3) cells | |
| Peptide, recombinant protein | AncIpCS1 | This paper | Purified from E. coli BL21(DE3) cells | |
| Peptide, recombinant protein | AncIpCS2 | This paper | Purified from E. coli BL21(DE3) cells | |
| Commercial assay or kit | DNeasy Plant Mini Kit | QIAGEN | Cat. #: 69104 | Used to extract genomic DNA from Ilex paraguariensis |
| Commercial assay or kit | Quick-DNA HMW MagBead Kit | Zymo Research | Cat. #: D6060 | Used to extract genomic DNA from Ilex paraguariensis |
| Commercial assay or kit | Illumina TruSeq DNA Sample Preparation Kit | Illumina | Cat. #: FC-121-2003 | Used to construct paired-end libraries |
| Commercial assay or kit | Illumina Nextera Mate Pair Library Preparation Kit | Illumina | Cat. #: FC-132-1001 | Used to construct mate-pair libraries |
| Commercial assay or kit | Sequel Binding Kit 1.0 | Pacific Biosciences | Cat. #: 101-365-900 | Used for preparing DNA templates for sequencing on the PacBio Sequel System |
| Commercial assay or kit | Sequel Sequencing Kit 1.0 | Pacific Biosciences | Cat. #: 101-309-500 | Used to perform sequencing reactions on the PacBio Sequel System |
| Commercial assay or kit | SMRT Cell 1M | Pacific Biosciences | Cat. #: 100-171-800 | Consumable microchip used in the PacBio Sequel System for Single Molecule, Real-Time (SMRT) sequencing |
| Commercial assay or kit | pTrcHis TOPO TA Expression Kit | Invitrogen | Cat. #: K4410-01 | Used to clone IpCS2 gene |
| Commercial assay or kit | QIAEX II Gel Extraction Kit | QIAGEN | Cat. #: 20021 | Used to clone IpCS1, IpCS3, AncIpCS1, and AncIpCS2 genes into pET-15b expression vector |
| Commercial assay or kit | Agilent QuikChange Lightning Kit | Agilent Technologies Inc, Santa Clara, CA | Cat. #: 210518 | Used for site-directed mutagenesis of AncIpCS2 |
| Commercial assay or kit | QIAprep Spin Miniprep Kit | QIAGEN | Cat. #: 27104 | Used for the rapid purification of high-quality plasmid DNA |
| Commercial assay or kit | TALON spin columns | Takara Bio | Cat. #: 89068 | Used for the purification of histidine-tagged proteins |
| Chemical compound, drug | Xanthine | Sigma-Aldrich | Cat. #: X0626 | Used to test relative substrate preference of IpCS1–3 and AncIpCS1–2 |
| Chemical compound, drug | Xanthosine | Sigma-Aldrich | Cat. #: X0750 | Used to test relative substrate preference of IpCS1–3 and AncIpCS1–2 |
| Chemical compound, drug | 1-Methylxanthine | Sigma-Aldrich | Cat. #: 69720 | Used to test relative substrate preference of IpCS1–3 and AncIpCS1–2 |
| Chemical compound, drug | 3-Methylxanthine | Sigma-Aldrich | Cat. #: 222526 | Used to test relative substrate preference of IpCS1–3 and AncIpCS1–2 |
| chemical compound, drug | 7-Methylxanthine | Sigma-Aldrich | Cat. #: 69723 | Used to test relative substrate preference of IpCS1–3 and AncIpCS1–2 |
| Chemical compound, drug | Theobromine | Sigma-Aldrich | Cat. #: T4500 | Used to test relative substrate preference of IpCS1–3 and AncIpCS1–2 |
| Chemical compound, drug | Paraxanthine | Sigma-Aldrich | Cat. #: D5385 | Used to test relative substrate preference of IpCS1–3 and AncIpCS1–2 |
| Chemical compound, drug | Theophylline | Sigma-Aldrich | Cat. #: T1633 | Used to test relative substrate preference of IpCS1–3 and AncIpCS1–2 |
| Software, algorithm | Trimmomatic | DOI: 10.1093/bioinformatics/btu170 | v.0.39 | Used to remove adaptor contaminations and filter low-quality reads |
| Software, algorithm | Quake | DOI: 10.1186/gb-2010-11-11-r116 | v.0.3 | Used to correct clean reads |
| Software, algorithm | SOAPdenovo | DOI: 10.1186/2047-217X-1-18 | v.2 | Used to assemble and scaffold contigs |
| Software, algorithm | DeconSeq | DOI: 10.1371/journal.pone.0017288 | v.0.4.3 | Used to detect and remove sequence contaminants |
| Software, algorithm | Canu | DOI: 10.1101/gr.215087.116 | v.2.2 | Used for self-correction and assembly of long reads |
| Software, algorithm | PurgeHaplotigs | DOI: 10.1186/s12859-018-2441-2 | Used to separate assembly haplotypes | |
| Software, algorithm | Quickmerge | DOI: 10.1101/029306 | v.03 | Used to merge SOAPdenovo and Canu curated assemblies |
| Software, algorithm | SSPACE | DOI: 10.1093/bioinformatics/btq683 | v.2.1.1 | Used to refine scaffolds and contigs |
| Software, algorithm | RepeatMasker | http://repeatmasker.org/ | Used to mask the genome assembly | |
| Software, algorithm | Funannotate | DOI: 10.5281/zenodo.2604804 | v.1.8.13 | Used to predict the protein- and non-coding genes |
| Software, algorithm | Infernal | DOI: 10.1093/bioinformatics/btt509 | v.1.1.4 | Used to improve the prediction of small RNAs and microRNAs |
| Software, algorithm | tRNAScan-SE | DOI: 10.1007/978-1-4939-9173-0_1 | v.2.0 | Used to improve the prediction of transfer RNAs |
| software, algorithm | TAPIR | http://bioinformatics.psb.ugent.be/webtools/tapir | Used to identify miRNA targets | |
| Software, algorithm | TargetFinder | DOI: 10.1007/978-1-60327-005-2_4 | v.1.7 | Used to identify miRNA targets |
| Software, algorithm | InterProScan | DOI: 10.1093/bioinformatics/btu031 | v.5.55-88.0 | Used to assign function of the predicted genes |
| Software, algorithm | eggNOG-mapper | DOI: 10.1093/nar/gky1085 | v.2.1.7 | Used to assign function to the predicted genes |
| Software, algorithm | Dfam TE Tools | https://github.com/Dfam-consortium/TETools | v.1.5 | Used to estimate the repeat content |
| Software, algorithm | CoGe’s tool SynMap | https://genomevolution.org/ | Used to estimate rates of synonymous substitution (Ks) between paralogous and orthologous genes | |
| Software, algorithm | CoGe's tool SynFind | https://genomevolution.org/ | Used to determine the syntenic depth ratio between I. paraguariensis, C. canephora, and V. vinifera | |
| Software, algorithm | CoGe’s tool GEvo | https://genomevolution.org/ | Used to compare CS and XMT syntenic regions | |
| Software, algorithm | MAFFT | DOI: 10.1093/molbev/mst010 | v.7.0 | Used to align amino acid sequences |
| Software, algorithm | FastTree | DOI: 10.1371/journal.pone.0009490 | v.2 | Used to perform phylogenetic analysis of SABATH sequences |
| Software, algorithm | IQTree | DOI: 10.1093/nar/gkw256 | Used to estimate ancestral sequences | |
| Software, algorithm | Phenix | DOI: 10.1107/S0907444909052925 | Used to solve the crystal structure of IpCS3 | |
| software, algorithm | REFMAC5 | DOI: 10.1107/S0907444911001314 | Used to refine the crystal structure of IpCS3 | |
| Software, algorithm | COOT | DOI: 10.1107/S0907444910007493 | v.0.9.8.3 | Used to refine the crystal structure of IpCS3 |
| Other | Ilex paraguariensis transcriptome sequence data | ENA | PRJNA315513 | Used to assess the completeness of Ilex paraguariensis genome |
| Other | Ilex paraguariensis transcriptome sequence data | NCBI | SRP043293 | Used to assess the completeness of Ilex paraguariensis genome |
| Other | Ilex paraguariensis transcriptome sequence data | NCBI | SRP110129 | Used to determine the expression of IpCS1–5 genes |
| Other | Vivaspin columns | Sartorius | Cat. #: VS0101 | Used to remove proteins after enzymatic reaction |
| Other | Kinetex 5 μM EVO C18 column | Phenomenex | Cat. #: 00F-4467-AN | Used for high-performance liquid chromatography |
| Other | Crystal Gryphon robot | Art Robbins Instruments | Cat. #: 100-1010 | Used for automating crystallization |





