Bromodomain inhibition of the transcriptional coactivators CBP/EP300 as a therapeutic strategy to target the IRF4 network in multiple myeloma
Figures
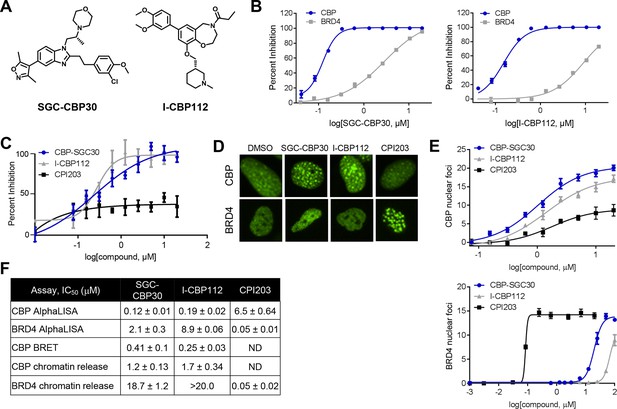
Characterization of CBP/EP300 bromodomain inhibitors.
(A) Structures of SGC-CBP30 and I-CBP112. (B) Representative AlphaLISA curves showing the inhibition of acetylated peptide binding to isolated CBP or BRD4 bromodomains in the presence of SGC-CBP30 and I-CBP112. Error bars represent SEM of 3 technical replicates. (C) Dose-titrations of SGC-CBP30, I-CBP112, and CPI203 using NanoBRET with the isolated CBP bromodomain and histone H3.3 in 293 cells. Error bars represent SEM of three technical replicates. The calculated EC50 values are shown in F. (D) ZsGreen-bromodomain fusion proteins were monitored by high content imaging. Representative nuclei showing nuclear foci in the indicated assays in the presence of DMSO, SGC-CBP30 (5 μM), I-CBP112 (5 μM) or CPI203 (0.33 μM). (E) Quantification of chromatin release assay. Each curve represents a titration of the indicated compound in stable cell lines expressing the indicated fusion protein (CBP: CBP-bromodomain/BRD9; BRD4: full length BRD4). Values are mean of four fields per well of two technical replicates, ± SEM. (F) Summary of biochemical and cellular activity of the indicated compounds. Values represent half-maximal inhibition (IC50) in AlphaLISA assays (n ≥ 2 independent replicates) or half-maximal induction (EC50) in NanoBRET (n = 3 technical replicates ± SEM) or chromatin release assays (n = 2 biological replicates ± SEM). ND = not determined due to a failure to produce 100% inhibition compared to controls.
-
Figure 1—source data 1
Bromodomain selectivity of CBP/EP300 bromodomain inhibitors.
Differential scanning fluorimetry was carried out with the indicated isolated bromodomains at 4–8 μM and the compounds at 20 μM. Shifts in melting temperature (△Tm, °C) and SEM for n = 3 technical replicates are shown.
- https://doi.org/10.7554/eLife.10483.004
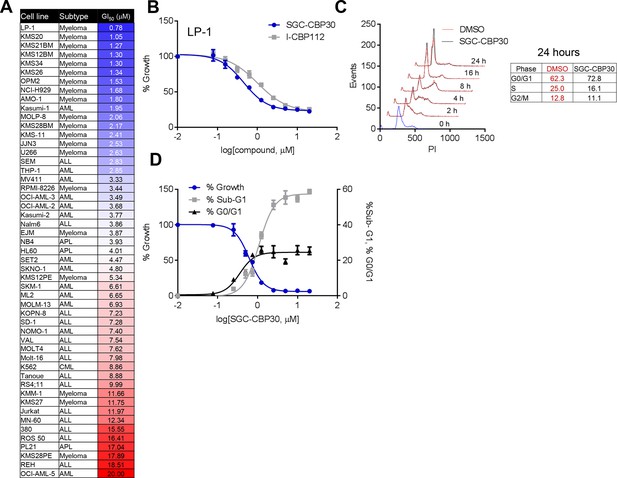
Phenotypic effects of CBP/EP300 bromodomain inhibition.
(A) Growth inhibitory effects of SGC-CBP30 and I-CBP112 in the indicated cell lines. Cells were incubated with compounds for 6 days, and viability was measured with resazurin. Values are the mean of at least two biological replicates. Values with error can be found in Figure 2—source data 1. (B) Example viability curves for LP-1. Values represent the mean of three3 technical replicates, ± SD. (C) LP-1 were synchronized by double thymidine block and released into either DMSO or 2.5 μM SGC-CBP30. Cells were fixed and stained with PI for cell cycle analysis at the indicated time points. Cell cycle distribution at 24 hr is shown in the table. Representative data from one of two biological replicates are shown. (D) LP-1 cells were treated as in (A) and fixed after 6 days. Viable cell number and percent increase in G0/G1 or sub-G1 over DMSO were determined by flow cytometry. Each point is the mean of three technical replicates, ± SD. See Figure 2—figure supplement 1 for additional data with I-CBP112.
-
Figure 2—source data 1
GI50 and standard deviation for a minimum of two replicates for the data shown in Figure 2 and Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.10483.006
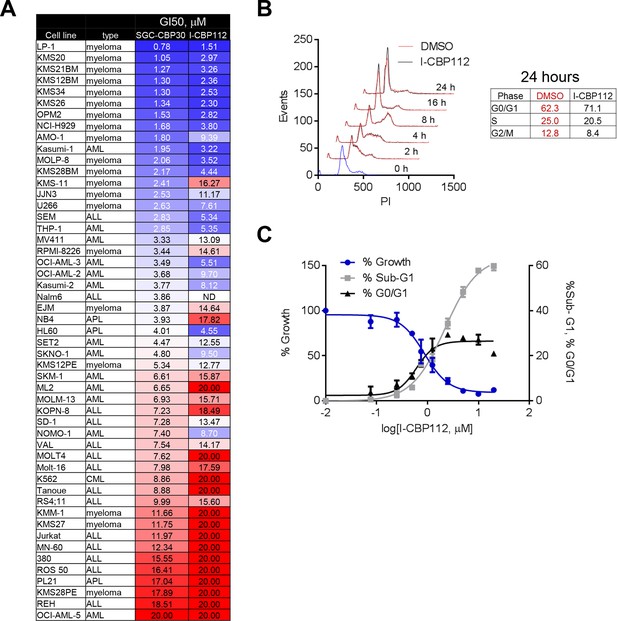
CBP/EP300 bromodomain inhibition affects the viability of multiple myeloma cells.
As in Figure 2, except with I-CBP112. (A) as in Figure 2A. (B) as in Figure 2C. (C) as in Figure 2D.
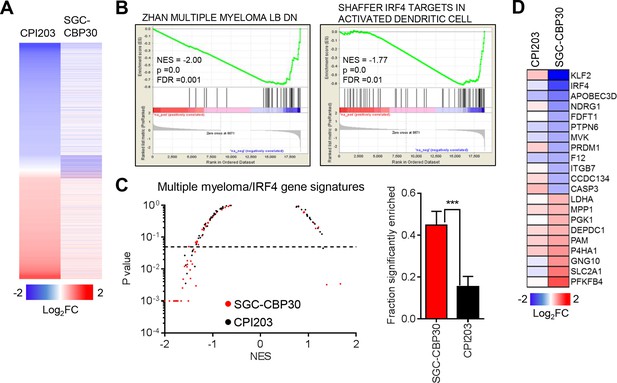
CBP/EP300 bromodomain inhibition targets IRF4.
(A) LP-1 cells were treated with SGC-CBP30 (2.5 μM) or CPI203 (0.25 μM) for 6 hr, and mRNA expression was measured using RNA sequencing. Expression values for biological replicate compound-treated samples were normalized to paired DMSO controls to obtain log2 fold change values. (B) Example enrichment plots for GSEA of SGC-CBP30 treated LP-1 cells. (C) Left, Scatter plot of P value vs. NES for multiple myeloma and IRF4 gene signatures for SGC-CBP30 (red) or CPI203 (black) treated LP-1 cells. Dashed line indicates p = 0.05. Right, fraction of gene signatures significantly enriched (p<0.05) with each treatment. Error bars indicate SEM. SGC-CBP30: 26/58; CPI203: 9/58. *** indicates p = 0.0005 by unpaired 2-tailed t-test. (D) IRF4 target genes differentially expressed (minimum 1.5 fold, p<0.05) with SGC-CBP30, but not CPI203. See Figure 3—figure supplement 1 for additional gene expression data and analysis.
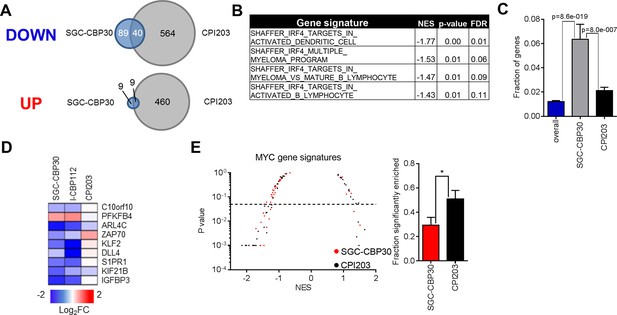
CBP/EP300 bromodomain inhibition targets IRF4 transcriptional programs.
(A) Venn diagrams showing the overlap of genes down- or up-regulated at least 2-fold following treatment with SGC-CBP30 or CPI203 as in Figure 3A. (B) Significantly enriched (p<0.05) IRF4 gene signatures upon SGC-CBP30 treatment. (C) The fraction of the 309 IRF4 target genes present in the overall set of mapped genes (20299 genes) or in the genes differentially expressed at least 1.5 fold by SGC-CBP30 (393 genes) or CPI203 (2959 genes) was determined. p-values were calculated by unpaired 2-tailed t-test. (D) Expression of the indicated mRNAs was determined by q-RTPCR following treatment of LP-1 cells with SGC-CBP30 (2.5 μM), I-CBP112 (5 μM), or CPI203 (0.25 μM) for 6 hr. Relative gene expression is expressed as log2 fold change relative to expression in DMSO. (E) Left, As in Figure 3C, except with MYC gene signatures. Right, fraction of gene signatures significantly enriched with each treatment. Error bars indicate SEM. SGC-CBP30: 15/51; CPI203: 26/51. * indicates p = 0.03 by unpaired 2-tailed t-test.
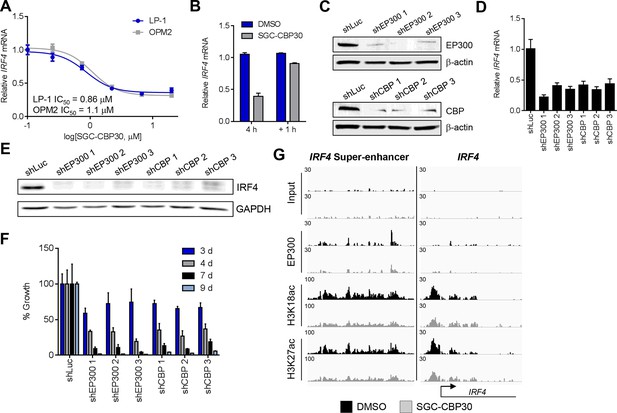
IRF4 is a direct transcriptional target of CBP/EP300 bromodomain inhibition.
(A) Dose-dependent inhibition of IRF4 mRNA expression (qRT-PCR) with SGC-CBP30 in LP-1 and OPM2 cells following 6 hr of treatment. Values represent the mean of three biological replicates, ± SEM. (B) LP-1 cells were treated with SGC-CBP30 (2.5 μM) for 4 hr, compound was removed, and cells were incubated for an additional 1 hr in fresh media. Levels of IRF4 mRNA were measured by qRT-PCR and normalized to GAPDH. Relative mRNA values normalized to DMSO at each time point represent the mean of 2 biological replicates, ± SEM. (C) Cells were transduced with lentivirus and lysed for Western analysis with the indicated antibodies (3 days post-infection). (D) IRF4 expression was determined by qRT-PCR at 3.5 days following the transduction of shRNA lentivirus, and mRNA was normalized to GAPDH and expressed relative to the control shLuc (n = 3 technical replicates, ± SEM). (E) Western analysis with the indicated antibodies was carried out at 3.5 days post-transduction with the indicated shRNA constructs. (F) Cells were fixed at the indicated time points following transduction and viability was determined by flow cytometry. Percent growth is expressed relative to control shLuc at each time point. Values represent the mean of n = 3 technical replicates, ± SEM. (G) LP-1 cells were treated with SGC-CBP30 (2.5 μM) for 6 hr, and the indicated antibodies were used for ChIP-seq. Sequencing traces for the IRF4 super-enhancer and the transcriptional start site are shown. See Figure 4—figure supplements 1 and 2 for additional supporting data.
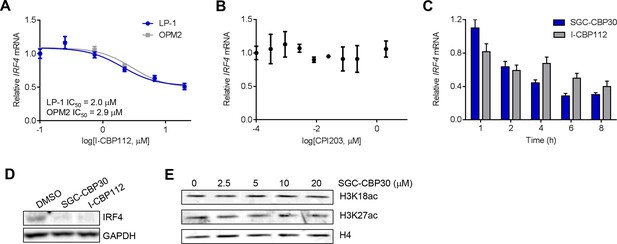
CBP/EP300 bromodomain inhibition regulates the expression of IRF4.
(A) Cells were treated with I-CBP112 for 6 hr, and levels of IRF4 were determined as in Figure 4A. Values represent the mean of n = 3 biological replicates ± SEM. (B) LP-1 cells were treated with a titration of CPI203 for 6 hr, and IRF4 expression was determined by qRT-PCR and normalized to GAPDH. Values represent the mean of n = 2 biological replicates, ± SEM. (C) LP-1 cells were treated with DMSO, SGC-CBP30 (2.5 μM), or I-CBP112 (5 μM). Total RNA was prepared at the indicated time points and used for qRT-PCR. Expression of IRF4 was normalized to GAPDH and calculated relative to DMSO treated cells at each time point. Values represent the mean of n = 4 technical replicates, ± SEM. (D) Uninduced LP-1/IRF4 cells were treated with SGC-CBP30 (2.5 μM) or I-CBP112 (5 μM) for 24 hr, and lysates were prepared for Western analysis with the indicated antibodies. (E) LP-1 cells were treated with the indicated concentrations of SGC-CBP30 for 16 hr, and extracts were prepared for Western analysis with the indicated antibodies.
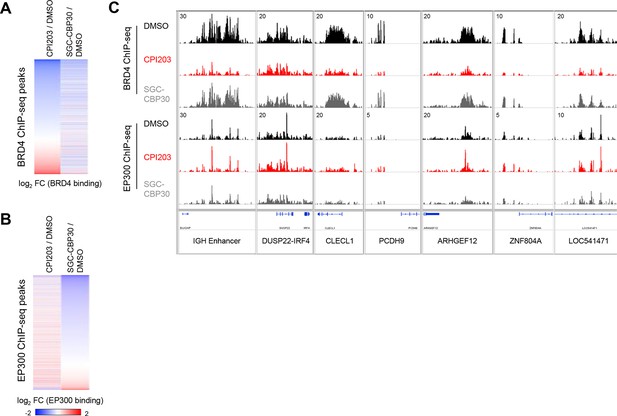
CBP/EP300 bromodomain inhibition does not cause global eviction of BRD4 from chromatin.
(A) BRD4 ChIP-seq peaks were called using MACS and ranked by log2 fold change in BRD4 enrichment in LP-1 cells treated for 6 hr with 0.25 µM CPI203 compared to DMSO-treated cells. (B) EP300 ChIP-seq peaks were called using MACS and ranked by log2 fold change in EP300 enrichment in LP-1 cells treated for 6 hr with 2.5 µM SGC-CBP30 compared to DMSO-treated cells. (C) Examples of BRD4 and EP300 ChIP-seq tracks showing that CBPi does not cause global eviction of BRD4, and that BETi does not globally reduce EP300 chromatin binding. Representative tracks of two biological replicates are shown.
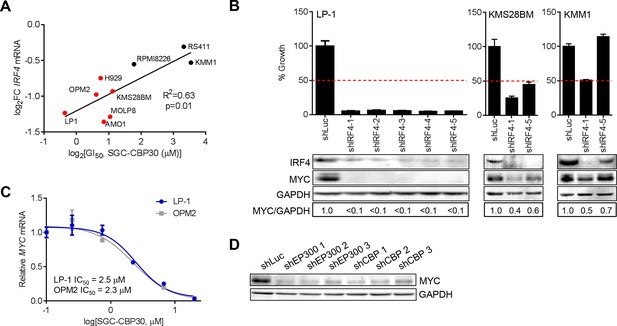
IRF4 suppression is correlated with phenotypic sensitivity to SGC-CBP30, and MYC is downregulated concomitant with IRF4 suppression following CBP/EP300 knockdown or bromodomain inhibition.
(A) The indicated cell lines were treated with SGC-CBP30 (2.5 μM) for 6 hr, and IRF4 expression normalized to GAPDH was determined by q-RTPCR. Suppression of IRF4 (log2 fold change relative to DMSO) was plotted against log2 GI50. R2 and p-value of the linear regression are shown. Cell lines indicated in red have a GI50 of less than 2.5 μM SGC-CBP30 (Figure 2A). Source data can be found in Figure 5—source data 1. (B) Lentiviral shRNA constructs were transduced into the indicated cell lines. Western analysis was carried out after 4 days, and viability (n = 3 technical replicates ± SEM), was assessed after 7 days. Intensity of MYC bands relative to GAPDH bands is shown below the Western blots. (C) Cells were treated as in Figure 4A, and normalized expression of MYC was determined by q-RTPCR. Values represent the mean of three biological replicates, ± SEM. (D) LP-1 cells were transduced as in Figure 4E, and MYC protein expression was determined by Western analysis. See Figure 5—figure supplements 1 and Figure 5—source data 1 for additional data.
-
Figure 5—source data 1
Source data for Figure 5A and Figure 5—figure supplement 1B.
- https://doi.org/10.7554/eLife.10483.014
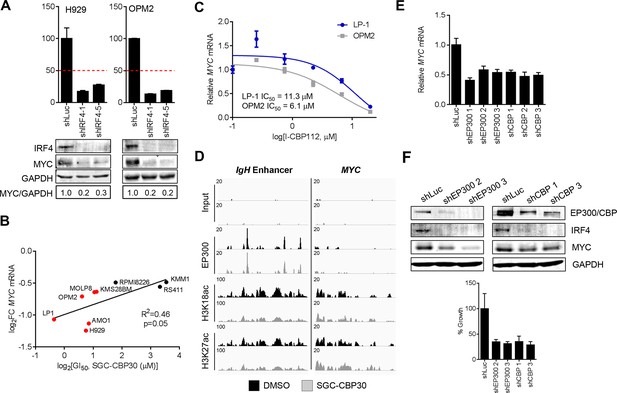
Suppression of the IRF4/MYC axis is important for the effects of CBP/EP300 bromodomain inhibition.
(A) The indicated cell lines were transduced as in Figure 5B, and Western analysis and viability were assessed as in Figure 5B. (B) As in Figure 5A, except with MYC expression. (C) Cells were treated with I-CBP112 for 6 hr, and levels of MYC were determined as in Figure 4A. Values represent the mean of n = 3 biological replicates ± SEM. (D) Cells were treated as in Figure 4G, and sequencing traces at the IgH enhancer and the MYC transcriptional start site are shown. (E) As in Figure 4D, except with MYC expression. Values represent the mean of n = 3 technical replicates, ± SEM. (F) OPM2 cells were transduced with the indicated shRNAs. Western analysis was carried out after 4 days, and viability was assessed by flow cytometry after 7 days. Values represent the mean of n = 3 technical replicates, ± SEM.
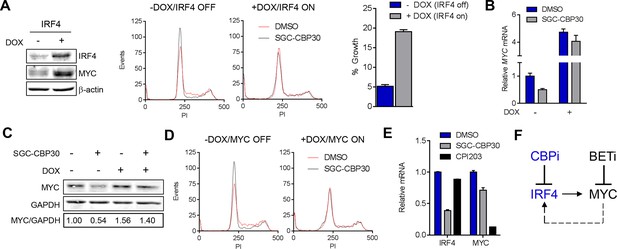
CBP/EP300 bromodomain inhibition suppresses the IRF4/MYC axis to cause viability defects.
(A) IRF4 expression was induced in the LP1/IRF4 cell line by the addition of doxycycline. Left, lysates were prepared after 3 days and used for Western analysis with the indicated antibodies. Middle, cells were incubated for an additional 24 hr with DMSO or SGC-CBP30 (2.5 μM) and fixed for cell cycle analysis by flow cytometry. Representative histograms of two biological replicate experiments are shown. Right, Cells were incubated for 6 days in the presence of SGC-CBP30 (2.5 μM). Viable cells were counted by flow cytometry and percent growth was calculated relative to the DMSO-treated condition for induced or uninduced cells. Values represent the mean of n = 3 technical replicates, ± SEM (B) Cells were induced as in (A) and were treated with DMSO or SGC-CBP30 (2.5 μM) for 6 hr. Expression of MYC was measured by qRT-PCR, normalized to GAPDH, and expressed relative to uninduced cells treated with DMSO. Values represent the mean of n = 3 technical replicates, ± SEM. (C) As in (B) except cells were treated for 24 hr and lysed for Western analysis with the indicated antibodies. Values represent the ratio of GAPDH-normalized MYC expression relative to uninduced DMSO-treated cells. (D) MYC expression was induced in the LP1/MYC cell line by the addition of doxycycline. Cells were incubated for an additional 24 hr with DMSO or SGC-CBP30 (2.5 μM) and fixed for cell cycle analysis by flow cytometry. Representative histograms of two independent experiments are shown. (E) RNA sequencing data from Figure 3A is expressed as the mean of the two biological replicates ( ± SEM) normalized to DMSO-treated cells. (F) Model for the suppression of the IRF4/MYC axis by CBP/EP300 and BET bromodomain inhibitors.
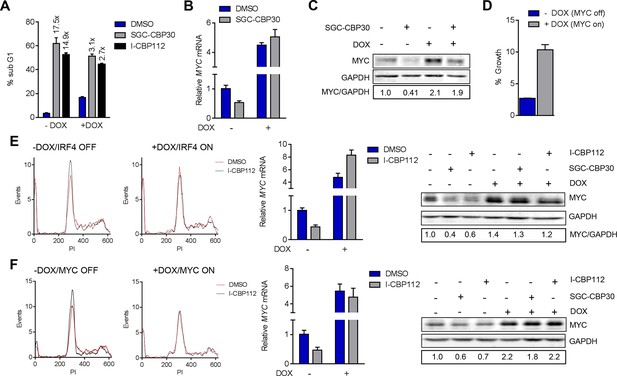
Additional data pertaining to IRF4 and MYC reconstitution experiments in Figure 6.
(A) Quantification of % sub G1 following 7 d of treatment with the DMSO, SGC-CBP30 (2.5 μM), or I-CBP112 (5 μM) in the absence (-DOX) or presence (+DOX) of ectopic IRF4. Fold increase above DMSO treatment for each condition is shown above the bars. Values represent the mean and SEM of n = 3 technical replicates. (B, C, and D,) as in Figure 6A,B, and C, except with the LP-1/MYC cell line. E and F as in Figure 6, except with I-CBP112 at 5 μM.
Additional files
-
Supplementary file 1
Includes qPCR primer sequences and UPL probe numbers for RT-qPCR experiments described in the manuscript.
- https://doi.org/10.7554/eLife.10483.018
-
Supplementary file 2
Indicates the source of all cell lines used, as well as results of mycoplasma testing throughout the course of these studies.
- https://doi.org/10.7554/eLife.10483.019





