Human eIF2A has a minimal role in translation initiation and in uORF-mediated translational control in HeLa cells
Figures
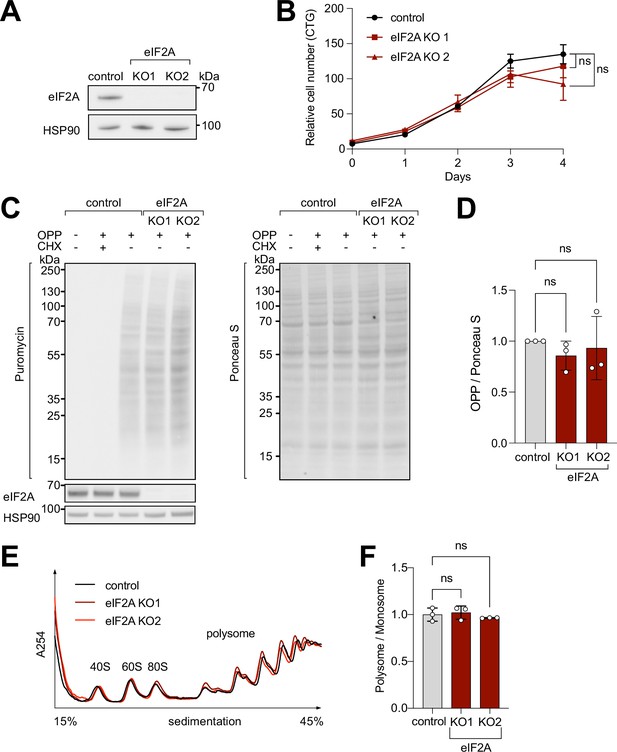
eIF2A has minimum effect on cell proliferation and global translation.
(A) Validation that eIF2A knockout cells have no eIF2A protein by immunoblotting. (B) Two independent eIF2A knockout HeLa cell lines have no proliferation defect, assayed by CellTiter Glo. Error bars: standard deviation. Significance by ordinary one-way ANOVA. (C–D) eIF2A knockout HeLa cells have no detectable change in global translation rates compared to control cells. (C) The translation rate was measured by immunoblotting to detect O-propargyl-puromycin (OPP) incorporated by metabolic labeling (left), and normalized to total protein amount assayed by Ponceau S (right). Three independent replicates are quantified in panel (D). CHX = cycloheximide treated sample to completely block global translation (negative control). Error bars: standard deviation. Significance by ordinary one-way ANOVA. (E–F) Polysome profiles of eIF2AKO cells show little to no difference to profiles from control cells. Lysates from either control or eIF2AKO HeLa cells were separated on a sucrose gradient. One representative graph is shown in panel E. The polysome/80 S ratio of three independent replicates is shown in panel F. Error bars: standard deviation. Significance by ordinary one-way ANOVA.
-
Figure 1—source data 1
pdf: Uncropped western blots.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig1-data1-v1.pdf
-
Figure 1—source data 2
Original immunoblot files from Chemidoc.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig1-data2-v1.zip
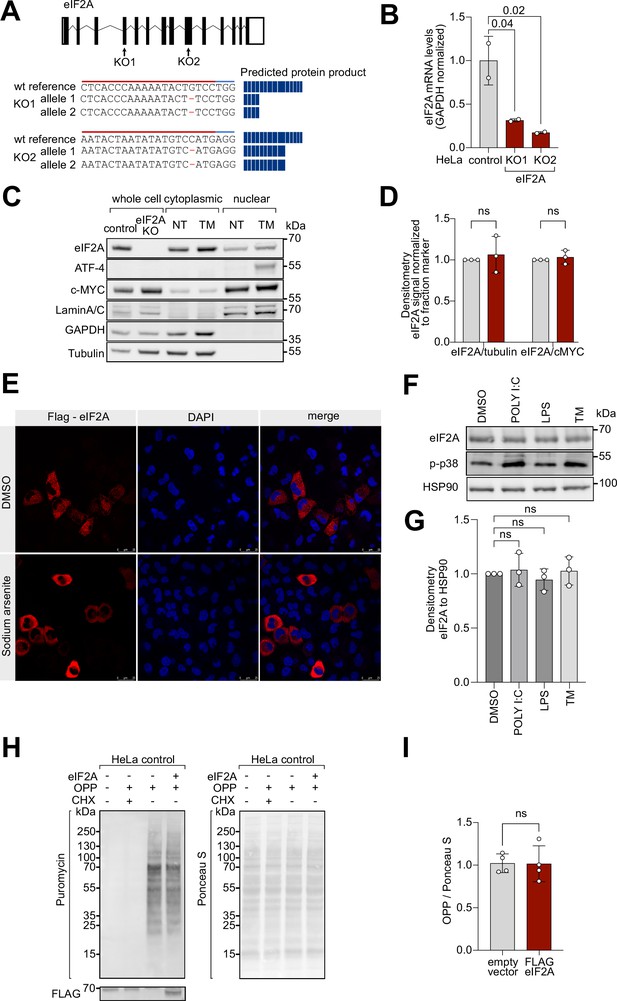
Loss of eIF2A does not perturb cell proliferation and global translation.
(A) Genotyping of eIF2AKO cell lines. Two eIF2A knockout lines were generated by targeting two separate coding exons. Shown is the resulting mutation at the DNA level, and a schematic of the predicted protein product. (B) mRNA levels of eIF2A, quantified by qRT-PCR, are reduced in the eIF2AKO lines. Error bars: standard deviation. Significance by ANOVA with Dunnett’s multiple comparison test. (C–D) Subcellular localization of eIF2A, detected by immunoblotting cytosolic and nuclear fractions or whole cell lysate (WCL), does not change upon treatment with tunicamycin (TM). Cells were treated with DMSO or Tunicamycin (1 µg/ml) for 2 hr. (C). Representative immunoblot (C) of three independent replicates quantified in (D). Nuclear and cytosolic fractions were lysed in equal volumes to reflect the relative contribution of the two compartments in a cell. Error bars: deviation. Significance by multiple unpaired, t-test. (E) Overexpressed FLAG-eIF2A shows predominantly cytoplasmic localization, assessed by immunofluorescent staining with anti-FLAG antibodies. Cells transfected with plasmid expressing eIF2A-FLAG were treated for 1 hr either with DMSO or 100 µM Sodium arsenite (SA). (F–G) eIF2A levels in HeLa cells are not affected by poly (I:C) (1 µg/ml), LPC (1 µg/ml), or tunicamycin (1 µg/ml) for 3 hr. Phospho-p38 signal is used as a positive control for the treatment. Representative immunoblot (F) or three independent replicates quantified in (G). Error bars: standard deviation. Significance by ANOVA with Dunnett’s multiple comparison test. (H–I) Global protein translation levels in HeLa cells do not change upon Flag-eIF2A overexpression, assessed by immunoblot against OPP (left) and normalized to total protein amount assayed by Ponceau S (right). Representative blots in (H), of four independent replicates quantified in (I). Error bars: standard deviation. Significance by unpaired, two-sided, t-test. All panels: ns = not significant.
-
Figure 1—figure supplement 1—source data 1
pdf: Uncropped western blots.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig1-figsupp1-data1-v1.pdf
-
Figure 1—figure supplement 1—source data 2
pdf: Uncropped western blots.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig1-figsupp1-data2-v1.pdf
-
Figure 1—figure supplement 1—source data 3
Original immunoblot files from Chemidoc.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig1-figsupp1-data3-v1.zip
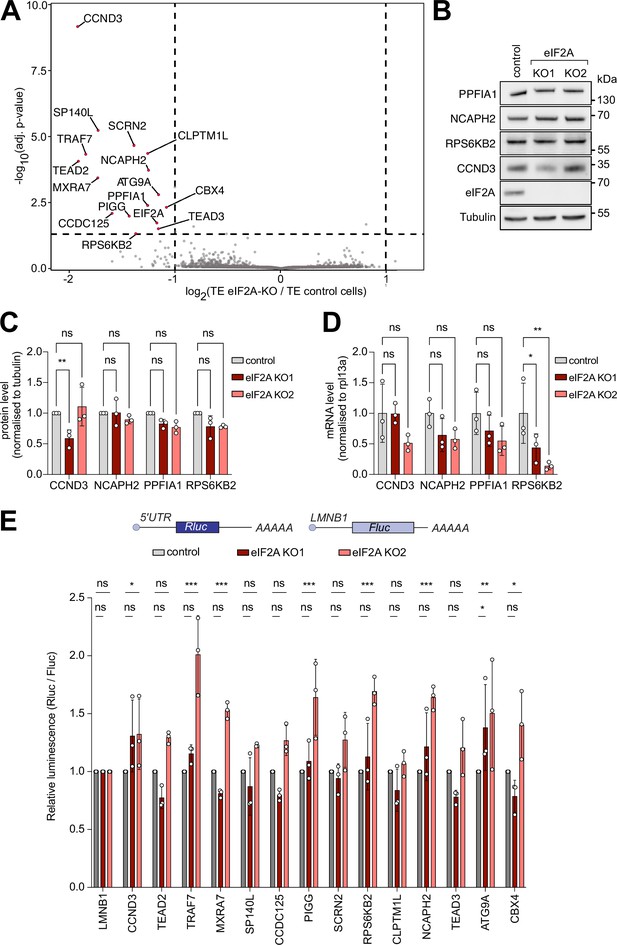
Ribosome profiling of eIF2A-KO lines finds little impact of eIF2A on translation.
(A) Ribosome profiling identifies a handful of mRNAs sensitive to eIF2A depletion. Scatter plot of log2(fold change of Translation Efficiency eIF2AKO/control) versus significance. Significant candidates with log2(fold change) < –1 are shown in red. Significance was estimated with the Wald test performed by the DESeq2 package. p-values are adjusted for multiple comparisons.(B–D) Western blot validation of ribosome profiling results. Among the tested candidates, only CCND3 shows decreased protein levels in one eIF2AKO clone. Representative blot in (B), of triplicates quantified in (C). mRNA levels of the corresponding transcripts are quantified and shown in (D). Significance by Dunnett’s multiple comparison test ANOVA. error bar = st. dev., ns = not significant, *p<0.05, **p<0.01. (E) Luciferase reporters harboring 5’ UTRs of eIF2A-dependent transcripts do not show strong changes in expression upon loss of eIF2A. Reporters carrying the 5’ UTRs of the indicated candidate genes were cloned upstream of Renilla Luciferase (RLuc) and co-transfected with a Firefly Luciferase (FLuc) normalization control. The negative control RLuc reporter and the FLuc normalization control carry the 5'UTR of Lamin B1 (LMNB1). Significance by Dunnett’s multiple comparison test ANOVA. error bar = st. dev., ns = not significant, *p<0.05, **p<0.01, ***p<0.001.
-
Figure 2—source data 1
pdf: Uncropped western blots.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig2-data1-v1.pdf
-
Figure 2—source data 2
Original immunoblot files from Chemidoc.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig2-data2-v1.zip
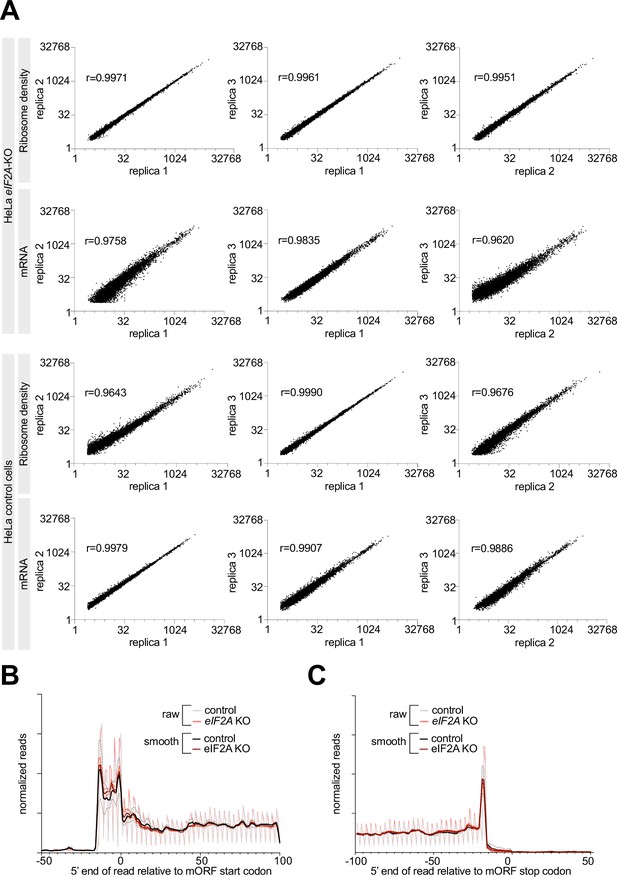
Ribosome profiling of control and eIF2AKO HeLa cells.
(A) Reproducibility between replicates of Ribosome profiling and total-mRNA libraries is shown. Three biological replicates were generated for control and eIF2AKO HeLa cells each. The Pearson’s coefficient (r) is shown for each compared pair. (B–C) Metagene profiles of footprints aligned to either the start codon (B) or the stop codon (C) of all main Open Reading Frames, for control and eIF2AKO HeLa cells. ‘Smooth’ curves were generated by averaging read counts with the sliding window of 3 nt. The dotted lines indicated the standard deviation between three replicates.
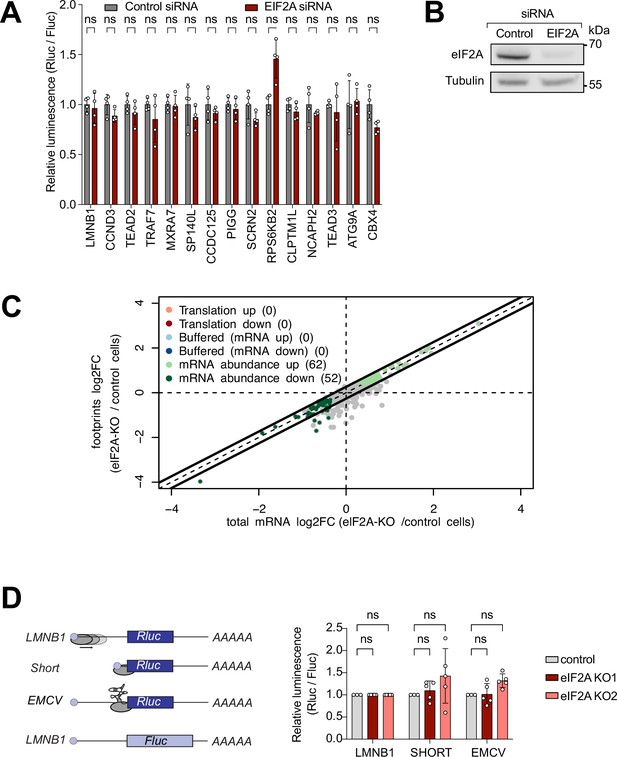
Loss of eIF2A does not affect translation of multiple different types of reporters.
(A) Luciferase reporters harboring 5’ UTRs of transcripts predicted to be eIF2A-dependent from ribosome footprinting do not show significantly reduced translation upon siRNA-mediated knockdown of eIF2A. The 5’ UTRs of the indicated genes were cloned upstream of Renilla Luciferase (RLuc) and co-transfected with a Firefly Luciferase (FLuc) normalization control reporter. Negative control RLuc reporter and the FLuc normalization control carry the 5'UTR of lamin B1 (LMNB1). Significance by multiple unpaired t-tests, ns = not significant. Error bars represent standard deviation. (B) Western blot control for efficiency of siRNA-mediated knockdown of eIF2A. (C) Transcripts shown in Figure 2A were reanalyzed with anota2seq package (Oertlin et al., 2019). Scatter plot of log2 fold-change of total RNA eIF2AKO/ control (x-axis) versus log2 fold-change of footprints eIF2AKO/ control (y-axis) is shown. Significant changes are detected in mRNA levels, while loss of eIF2A does not perturb translation. (D) Transfection of luciferase reporters designed to place the ribosome directly on top of the initiation AUG do not show reduced translation in eIF2AKO cells compared to controls. A 5’UTR containing the EMCV IRES or a short 5’ UTR of only 12 nt was cloned upstream of Renilla Luciferase (RLuc) and co-transfected with a FLuc normalization control. Negative control RLuc reporter and the FLuc normalization control carry the 5'UTR of Lamin B1 (LMNB1). Significance by ANOVA with Dunnett’s multiple comparison test. error bar = st. dev., ns = not significant.
-
Figure 2—figure supplement 2—source data 1
pdf: Uncropped western blots.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig2-figsupp2-data1-v1.pdf
-
Figure 2—figure supplement 2—source data 2
Original immunoblot files from Chemidoc.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig2-figsupp2-data2-v1.zip
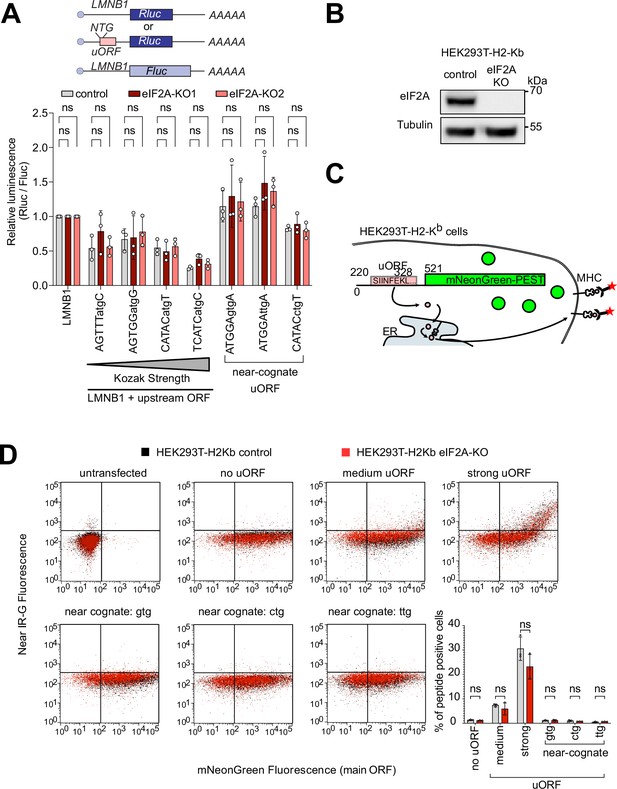
eIF2A has little or effect on uORF translation.
(A) Synthetic reporters harboring uORFs with different start codons and initiation contexts do not show dependence on eIF2A. The sequence context of the uORF start codons is indicated: either AUG or a near-cognate start codon (GTG, TTG, CTG) was used. Significance by Dunnett’s multiple comparison test ANOVA, error bar = st. dev. ns = not significant. (B) Validation that HEK293T-H2-Kb eIF2AKO cells have no eIF2A protein by immunoblotting. (C) Schematic diagram illustrating the setup to simultaneously detect a small peptide produced by a uORF and fluorescent mNeonGreen encoded by the main ORF. The short peptide SIINFEKL is presented on the cell surface by MHC-I and detected using a monoclonal antibody. (D) eIF2A knockout does not cause a drop in uORF translation. In the graph to the right, the percent of uORF-positive cells relative to all mNeonGreen cells is quantified. Significance by unpaired, two-sided, t-test. ns = not significant.
-
Figure 3—source data 1
pdf: Uncropped western blots.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig3-data1-v1.pdf
-
Figure 3—source data 2
Original immunoblot files from Chemidoc.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig3-data2-v1.zip
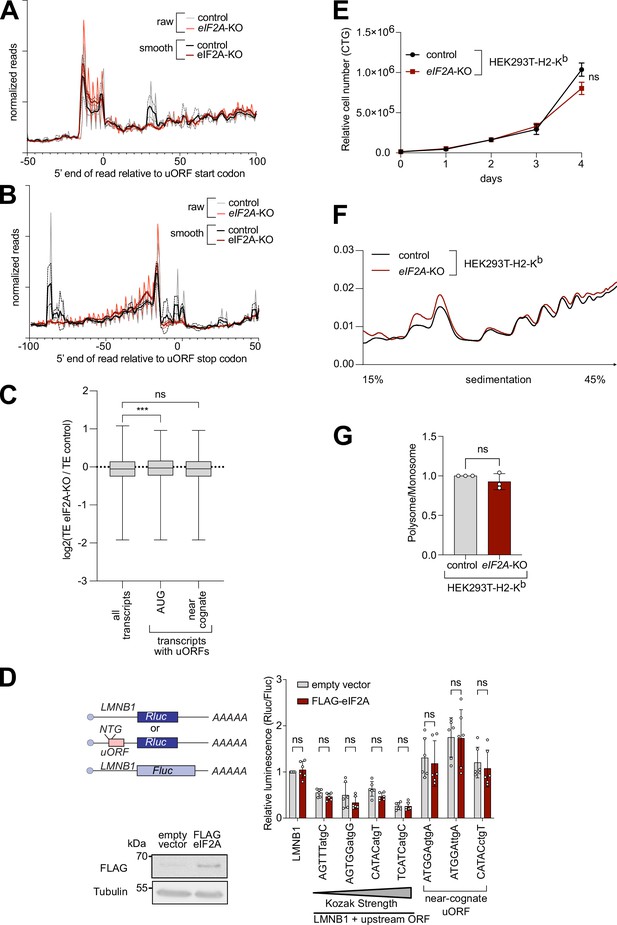
Knockout of eIF2A has no effect on uORF translation.
(A–B) Metagene profiles of footprints relative to either the start codon (A) or stop codon (B) of uORFs transcriptome-wide, for control or eIF2AKO HeLa cells. ‘Smooth’ curves were generated by averaging footprint counts with a sliding window of 3 nt. The dotted lines indicated the standard deviation between three replicates. (C) eIF2A has little impact genome-wide on translation of mRNAs with uORFs. Comparison of the change in translation efficiency between eIF2AKO and control HeLa cells for three different groups of mRNAs: all transcripts, transcripts with AUG-initiated uORFs, or transcripts with uORFs that start with a near-cognate codon. Significance by ANOVA with Dunnett’s multiple comparison test. ns = not significant, ***p<0.001. (D) Activity of synthetic reporters harboring uORFs with different start codons and initiation contexts is not altered upon eIF2A-overexpression. The sequence context of the uORF start codons is indicated: either AUG or a near-cognate start codon (GTG, TTG, CTG) was used. Overexpression of eIF2A was validated by western blot against the FLAG-tag. Significance by Dunnett’s multiple comparison test ANOVA, error bar = st. dev. ns = not significant. (E) Proliferation of eIF2AKO or control HEK293T-H2-Kb cells by CellTiter Glo. Error bars: standard deviation. Significance by unpaired, two-sided, t-test. ns = not significant. (F–G) Global cellular translation, assayed via polysome profiles, shows little difference between eIF2AKO and control HEK293T-H2-Kb cells. Lysates from either eIF2AKO or control HEK293T-H2-Kb cells were separated on sucrose gradients. One representative graph is shown in (F). The polysome/80 S ratio of three independent replicates is shown in (G). Error bars: standard deviation. Significance by unpaired, two-sided, t-test. ns = not significant.
-
Figure 3—figure supplement 1—source data 1
pdf: Uncropped western blots.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig3-figsupp1-data1-v1.pdf
-
Figure 3—figure supplement 1—source data 2
Original immunoblot files from Chemidoc.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig3-figsupp1-data2-v1.zip
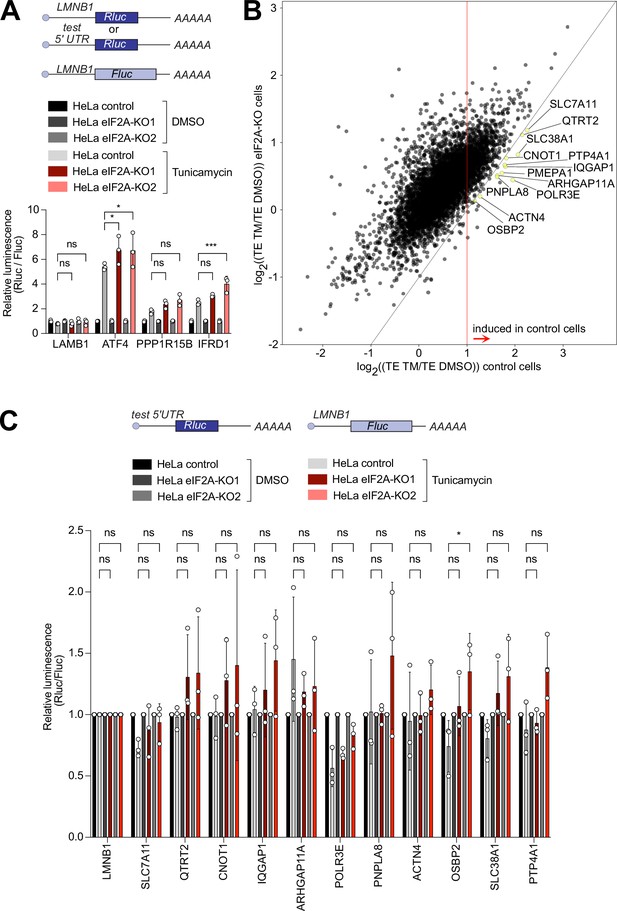
eIF2A has a minor impact on translation during the integrated stress response.
(A) Loss of eIF2A does not blunt induction of target genes of the integrated stress response. Reporters carrying the 5’ UTRs of the indicated candidate genes were co-transfected with a Firefly Luciferase (FLuc) normalization control reporter into eIF2AKO and control HeLa cells and treated for 16 hr either with DMSO or 1 µg/ml tunicamycin (TM). Significance by Dunnett’s multiple comparison test ANOVA. error bar = st. dev., ns = not significant, *p<0.05, ***p<0.001 (B) Ribosome profiling identifies 12 mRNAs that are significantly induced upon tunicamycin treatment (1 ug/ml) in control cells but not eIF2AKO cells. Scatter plot of log2(fold change) of Translation Efficiency TM/DMSO for control cells on the x-axis versus eIF2AKO cells on the y-axis. mRNAs that are statistically significantly induced with log2(fold change)>1 in control cells but not in eIF2AKO cells are shown in yellow and marked by gene name. Significance was estimated with the Wald test performed by DESeq2 thepackage. p-values are adjusted for multiple comparisons. (C) Transfection of luciferase reporters harboring 5’ UTRs of eIF2A-dependent transcripts does not show impaired induction in eIF2AKO cells upon tunicamycin treatment. 5’ UTRs of eIF2A-dependent transcripts from panel B were cloned upstream of Renilla luciferase and co-transfected with a FLuc normalization control reporter into control or EIF2AKO HeLa cells with subsequent treatment for 16 hr either with DMSO or 1 μg/ml TM. Significance by Dunnett’s multiple comparison test ANOVA. error bar = st. dev., ns = not significant, *p<0.05.
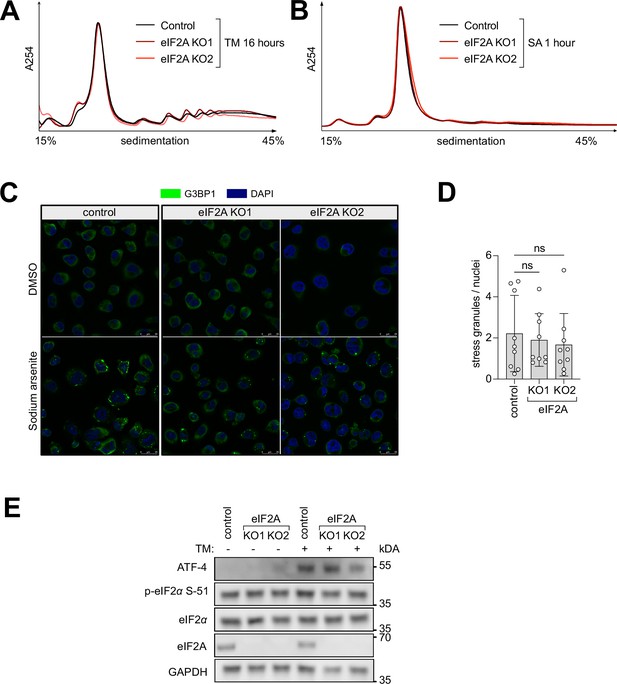
The integrated stress response suppresses translation equally well in control and eIF2AKO HeLa cells.
(A–B) Polysome profiles of eIF2AKO versus control HeLa cell lines show hardly any differences upon stress caused either with (A) 1 µg/ml tunicamycin (TM) for 16 hr or (B) 100 µM sodium arsenite (SA) for 1 hr. Profiles were aligned by the height of the 80 S peak. (C–D) eIF2A knockout cells form stress granules to the same degree as the parental control HeLa cell line. Control or eIF2AKO HeLa cells were treated for 2 hr with 100 µM sodium arsenite and stained for G3BP1. (C) Representative images. (D) The number of stress granules, normalized to the number of nuclei in each field of view is shown. Significance by ANOVA with Dunnett’s multiple comparison test. ns = not significant. (E) eIF2AKO and control HeLa cells phosphorylate eIF2α to the same degree in response to tunicamycin (1 µg/mL for 16 hr). ATF-4 is used as a marker for downstream activation of the integrated stress response.
-
Figure 4—figure supplement 1—source data 1
pdf: Uncropped western blots.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig4-figsupp1-data1-v1.pdf
-
Figure 4—figure supplement 1—source data 2
Original immunoblot files from Chemidoc.
- https://cdn.elifesciences.org/articles/105311/elife-105311-fig4-figsupp1-data2-v1.zip
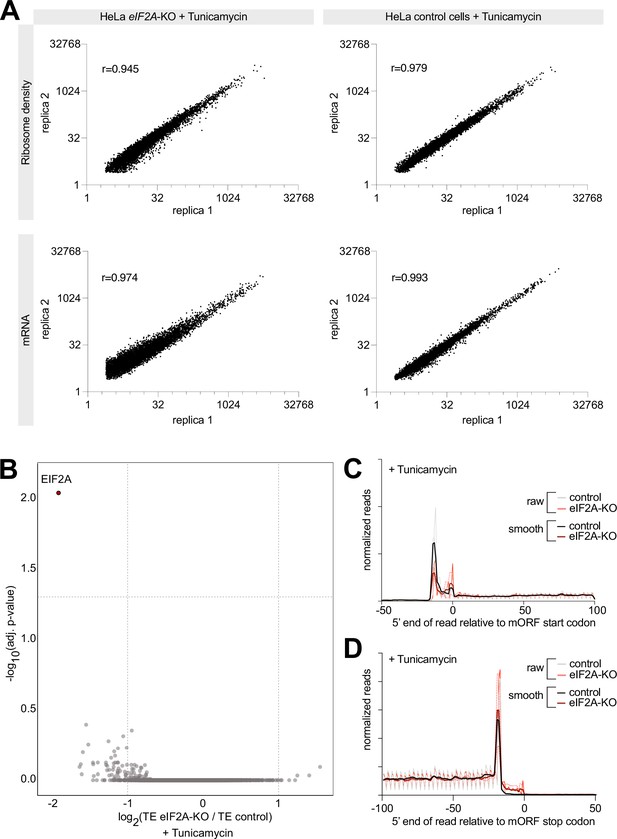
Ribosome profiling of tunicamycin-treated eIF2AKO and control HeLa cells.
(A) Reproducibility between replicates of Ribosome profiling or total-mRNA libraries from eIF2AKO or control HeLa cells treated with tunicamycin. Two biological replicates were generated for each genotype. The Pearson’s coefficient (r) is shown for each comparison. (B) Ribosome profiling identifies zero transcripts affected by eIF2A knockout in tunicamycin-treated conditions. Scatter plot of log2(fold change) of Translation Efficiency comparing eIF2AKO to control HeLa cells, both treated with tunicamycin, on the x-axis, versus significance on the y-axis. Significant candidates with log2(fold change) < –1 are shown in red. Significance was estimated with the Wald test performed by the DESeq2 package. p-values are adjusted for multiple comparisons. (C–D) Metagene profiles of footprints aligned to either the start codon (C) or the stop codon (D) of all main Open Reading Frames, for control and eIF2AKO HeLa cells, both treated with tunicamycin. ‘Smooth’ curves were generated by averaging read counts with the sliding window of 3 nt. The dotted lines indicate standard deviation between three replicates.
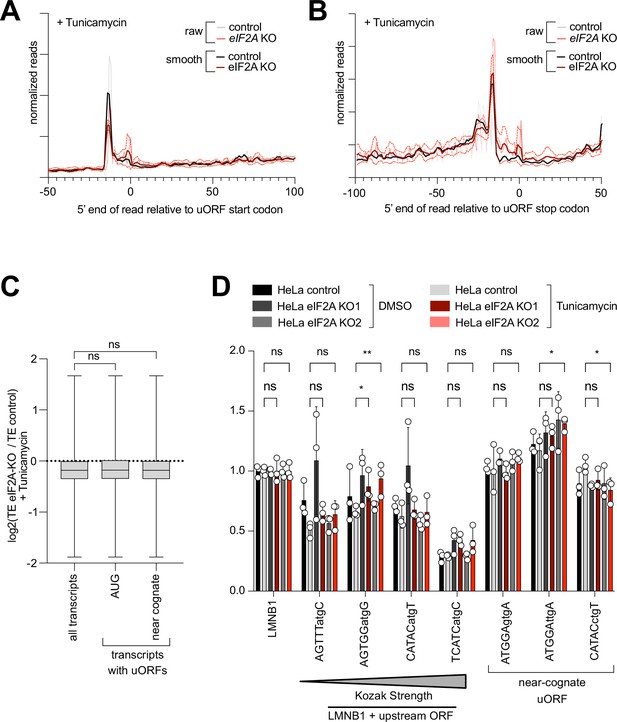
Translation of uORF-bearing transcripts is not affected upon loss of eIF2A in tunicamycin-treated cells.
(A–B) Metagene profiles of footprints relative to either the start codon (A) or stop codon (B) of uORFs transcriptome-wide, for control or eIF2AKO HeLa cells, both treated with tunicamycin. ‘Smooth’ curves were generated by averaging footprint counts with a sliding window of 3 nt. The dotted lines indicated the standard deviation between three replicates. (C) eIF2A has little impact genome-wide on translation of mRNAs with uORFs in cells treated with tunicamycin. Comparison of the change in translation efficiency between eIF2AKO and control HeLa cells, both treated with tunicamycin, for three different groups of mRNAs: all transcripts, transcripts with AUG-initiated uORFs, or transcripts with uORFs that start with a near-cognate codon. Significance by ANOVA with Dunnett’s multiple comparison test. ns = not significant. (D) Synthetic reporters harboring uORFs with different start codons and initiation contexts do not show dependence on eIF2A also when the integrated stress response is activated, thereby suppressing eIF2 function. Either control or eIF2AKO HeLa cells were transfected with the indicated reporters and then treated with 1 µg/mL tunicamycin for 16 hr prior to assaying luciferase activity. The sequence context of the uORF start codons is indicated: either AUG or a near-cognate start codon (GTG, TTG, CTG) was used. Significance by Dunnett’s multiple comparison test ANOVA, error bar = st. dev. ns = not significant. *p<0.05, **p<0.01.
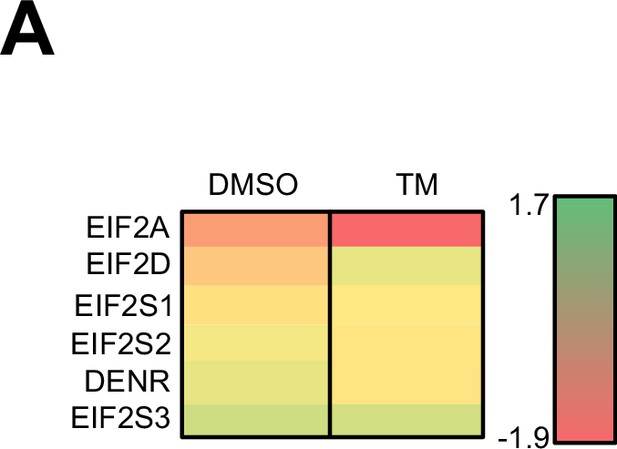
Expression of initiation factors reported to possess tRNAiMet binding activities.
(A) Heat map showing the log2 fold change of translation efficiency (eIF2AKO/control cells) of different initiation factors reported to possess binding capacity to tRNAiMet. Changes in DMSO and tunicamycin-treated samples are shown.
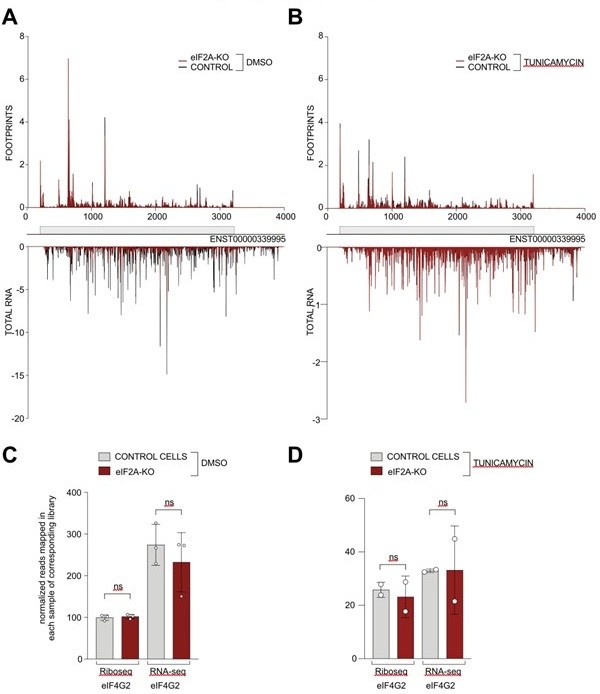
(A-B) Average reads occupancy on the eIF4G2 (ENST0000339995) transcript in DMSO treated (panel A, n=3) or tunicamycin treated samples (panel B, n=2) derived from either control (black) or eIF2A-KO (red) HeLa cells. Reads counts were normalized to sequencing depth and averaged between either 3 (DMSO-treated) or 2 (tunicamycin-treated) replicates. Graphs were then smoothened with a sliding window of 3 nt. (C-D) The total number of reads mapping to the eIF4G2 CDS, normalized to library sequencing depth per replica was quantified. No significant difference between control and eIF2A-KO cells was observed in either DMSO treated (panel C) or tunicamycin treated (panel D) cells. Significance by unpaired, two-sided, t-test. ns = not significant.
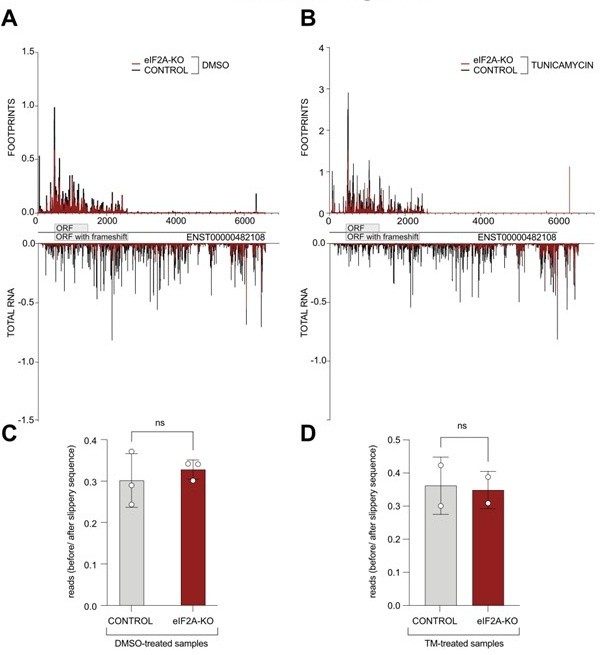
(A-B) Average reads occupancy on the PEG10 (ENST00000482108) transcript in DMSO treated (panel A, n=3) or tunicamycin treated samples (panel B, n=2) derived from either control (black) or eIF2A-KO (red) HeLa cells are shown. Reads counts were normalized to sequencing depth and averaged between either 3 (DMSO-treated) or 2 (tunicamycin-treated) replicates. Graphs were then smoothened with a sliding window of 3 nt. (C-D) The ratio of reads mapping to the ORF upstream of the slippery site to reads mapping to the predicted extended protein downstream to the slippery site is shown. Reads counts were normalized to the sequencing depth. Neither DMSO treated samples (panel C) nor tunicamycin treated samples (panel D) had a significant difference between control and eIF2A-KO cells. Significance by unpaired, two-sided, t-test. ns = not significant.
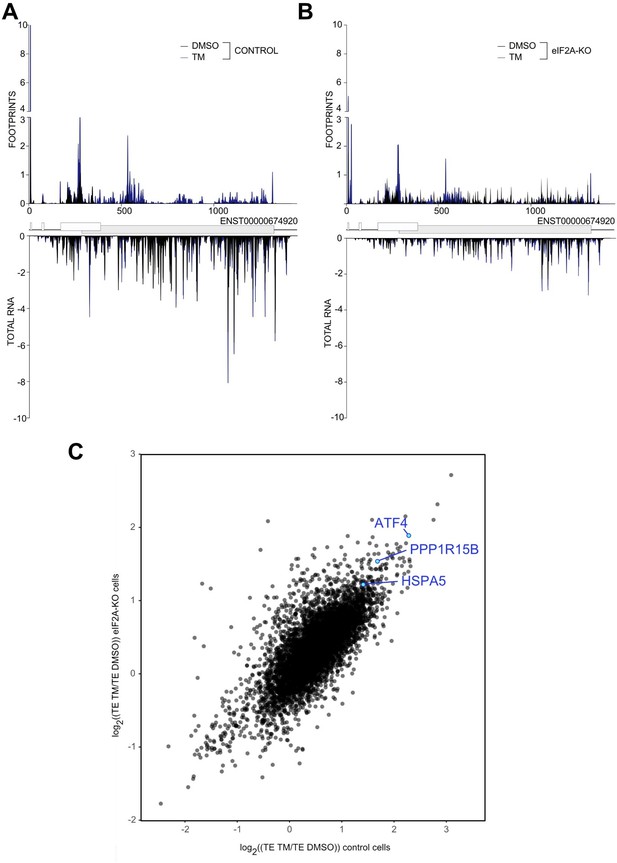
(A-B) Average read occupancy on the ATF4 (ENST00000674920) transcript in DMSO treated (n=3) or tunicamycin treated samples (n=2) derived from either control (panel A) or eIF2A-KO (panel B) HeLa cells are shown. Read counts were normalized to sequencing depth and averaged between either 3 (DMSO-treated) or 2 (tunicamycin-treated) replicates. Graphs were then smoothened with a sliding window of 3 nt. (C) Scatter plot of log2(fold change) of Translation Efficiency TM/DMSO for control cells on the xaxis versus eIF2AKO cells on the y-axis. The induction of ATF4 as well as the downstream target PPP1R15B are shown. The upregulation of HSP5A translation, the other hallmark of ER-stress induced by tunicamycin treatment is shown.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | eIF2A | Ensemble | ENSG00000144895 | |
| Cell line (Homo sapiens) | HeLa | DSMZ | ACC 57 | |
| Cell line (Homo sapiens) | HeLa-eIF2A-KO | This study | This study | |
| Cell line (Homo sapiens) | HEK293T-H2-Kb | gift from Rienk Offringa lab | Rienk Offringa lab | |
| Cell line (Homo sapiens) | HEK293T-H2-Kb-eIF2A-KO | This study | This study | |
| Transfected construct (human) | siRNA to eIF2A (mix of all 4 was used) | Horizon discovery | D-014766–01 | GCUCCCAGGUUACGGGUUA |
| Transfected construct (human) | D-014766–02 | GAUUUGGAAUUGGGUAUUU | ||
| Transfected construct (human) | D-014766–03 | GCAGAUAAAGUUACAAUGC | ||
| Transfected construct (human) | D-014766–04 | CCACAAUCAGGAAACGAUA | ||
| Sequence-based reagent (oligos used to produce KO) | Pair 8 sg_eIF2A 1 | This study | OMR064 | CACCGCTCACCCAAAAATACTGTCC |
| Sequence-based reagent (oligos used to produce KO) | Pair 8 sg_eIF2A 2 | This study | OMR065 | AAACGGACAGTATTTTTGGGTGAGC |
| Sequence-based reagent (oligos used to produce KO) | Pair 5 sg_eIF2A 1 | This study | OMR058 | CACCGAATACTAATATATGTCCATG |
| Sequence-based reagent (oligos used to produce KO) | Pair 5 sg_eIF2A 2 | This study | OMR059 | AAACCATGGACATATATTAGTATTC |
| Antibody | anti-ATF-4 (D4B8) (Rabbit monoclonal) | Cell Signaling | cat. No #11815 Lot#6 | WB (1:1000) |
| Antibody | anti-C-Myc (Rabbit monoclonal) | Cell Signaling | cat. No. #13987 Lot#6 | WB (1:1000) |
| Antibody | anti-CCND3 (Rabbit polyclonal) | invitrogen | cat no. #PA5-80416 Lot#UH2828593 | WB (1:1000) |
| Antibody | anti-eIF2A (3A7A8) (mouse monoclonal) | santa cruz | cat. No. sc-517214 Lot#B0821 | WB (1:1000) |
| Antibody | anti-FLAG (Rabbit polyclonal) | SIGMA | cat. No. F7425-.2MG Lot#0000252651 | WB (1:1000) |
| Antibody | anti-GAPDH (Rabbit monoclonal) | Cell Signaling | cat. No. #2118 LOT#16 | WB (1:1000) |
| Antibody | anti-HSP90 (Rabbit monoclonal) | Cell signaling | cat. No. 4877 Lot#6 | WB (1:1000) |
| Antibody | anti-Lamin A/C (636) (mouse monoclonal) | Santa Cruz | cat. No. sc-7292 Lot#C0218 | WB (1:1000) |
| Antibody | anti-NCAPH2 (Rabbit polyclonal) | Proteintech | cat. No. 26172–1-AP Lot#00039440 | WB (1:1000) |
| Antibody | anti-p-p38 (Rabbit polyclonal) | Cell Signaling | cat. No. #9211 Lot#25 | WB (1:1000) |
| Antibody | anti-PPFIA1 (Rabbit polyclonal) | Proteintech | cat. No. 14175–1-AP Lot#00005224 | WB (1:1000) |
| Antibody | anti-puromycin (mouse monoclonal) | Sigma | cat. No. MABE343 Lot#3484967 | WB (1:1000) |
| Antibody | anti-RPS6KB2 (Rabbit polyclonal) | Proteintech | cat. No. 26194–1-AP Lot#00040692 | WB (1:1000) |
| Antibody | anti-Tubulin | Sigma | cat. No. T9026 LOT#0000307925 | WB (1:2500) |
| Antibody | anti-G3BP1 (mouse monoclonal) | santa cruz | cat. No. sc-81940 Lot@G0617 | IF: 1:50 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | ACTN4 | This study | pMR1184 | 5' UTR cloned from transcript with id ENST00000252699 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | ARHGAP11A | This study | pMR181 | 5' UTR cloned from transcript with id ENST00000361627 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | ATG9A | This study | pMR091 | 5' UTR cloned from transcript with id ENST00000361242 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | CBX4 | This study | pMR1171 | 5' UTR cloned from transcript with id ENST00000269397 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | CCDC125 | This study | pMR1174 | 5' UTR cloned from transcript with id ENST00000383374 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | CCND3 | This study | pMR965 | 5' UTR cloned from transcript with id ENST00000372991 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | CLPTM1L | This study | pMR1178 | 5' UTR cloned from transcript with id ENST00000337392 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | CNOT1 | This study | pMR1182 | 5' UTR cloned from transcript with id ENST00000317147 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | IQGAP1 | This study | pMR168 | 5' UTR cloned from transcript with id ENST00000268182 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | MXRA7 | This study | pMR1173 | 5' UTR cloned from transcript with id ENST00000355797 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | NCAPH2 | This study | pMR1170 | 5' UTR cloned from transcript with id ENST00000420993 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | OSBP2 | This study | pMR664 | 5' UTR cloned from transcript with id ENST00000332585 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | PIGG | This study | pMR096 | 5' UTR cloned from transcript with id ENST00000310340 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | PNPLA8 | This study | pMR1183 | 5' UTR cloned from transcript with id ENST00000257694 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | POLR3E | This study | pMR1181 | 5' UTR cloned from transcript with id ENST00000640588 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | PTP4A1 | This study | pMR196 | 5' UTR cloned from transcript with id ENST00000626021 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | QTRT2 | This study | pMR1180 | 5' UTR cloned from transcript with id ENST00000281273 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | RPS6KB2 | This study | pMR1176 | 5' UTR cloned from transcript with id ENST00000312629 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | SCRN2 | This study | pMR1175 | 5' UTR cloned from transcript with id ENST00000290216 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | SLC38A1 | This study | pMR1185 | 5' UTR cloned from transcript with id ENST00000546893 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | SLC7A11 | This study | pMR1179 | 5' UTR cloned from transcript with id ENST00000280612 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | SP140L | This study | pMR1177 | 5' UTR cloned from transcript with id ENST00000415673 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | TEAD2 | This study | pMR1172 | 5' UTR cloned from transcript with id ENST00000311227 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | TEAD3 | This study | pMR1169 | 5' UTR cloned from transcript with id ENST00000338863 |
| Sequence-based reagent (plasmids with 5' UTR of interest cloned upstream of renilla luciferase) | TRAF7 | This study | pMR967 | 5' UTR cloned from transcript with id ENST00000326181 |
| Sequence-based reagent (qRT-PCR oligos) | eIF2A qRT-PCR | This study | OMR202 | AAAGCACAGTGTTTCCAAGGG |
| Sequence-based reagent (qRT-PCR oligos) | eIF2A qRT-PCR | This study | OMR203 | GCAGTAGTCCCTTGTTAGTGA |
| Sequence-based reagent (qRT-PCR oligos) | CCND3 qRT-PCR oligo | This study | OMR412 | GAAGGGGCGTCTGTTCC |
| Sequence-based reagent (qRT-PCR oligos) | CCND3 qRT-PCR oligo | This study | OMR413 | CAGGGAGGAGGAGCTTG |
| Sequence-based reagent (qRT-PCR oligos) | NCAPH2 qRT-PCR oligo | This study | OMR2542 | CGAGTATCTGGAGGAGCTGGATCA |
| Sequence-based reagent (qRT-PCR oligos) | NCAPH2 qRT-PCR oligo | This study | OMR2543 | GCCTGGTAGACGAGTGAGTAGAGG |
| Sequence-based reagent (qRT-PCR oligos) | PPFIA1 qRT-PCR oligo | This study | OMR2546 | AGCAGAAAGGAATAACACCAGGCT |
| Sequence-based reagent (qRT-PCR oligos) | PPFIA1 qRT-PCR oligo | This study | OMR2547 | CATCCAGAGCTTTGTGGTGTTCAA |
| Sequence-based reagent (qRT-PCR oligos) | RPS6KB2 qRT-PCR oligo | This study | OMR2550 | TGGATTTGGAGACGGAGGAAGGCA |
| Sequence-based reagent (qRT-PCR oligos) | RPS6KB2 qRT-PCR oligo | This study | OMR2551 | GATGCGCTCTGGGCCAACGTTCAC |
| Sequence-based reagent (qRT-PCR oligos) | RPL13A qRT-PCR oligo | This study | OMR068 | CCGCCCTACGACAAGAAA |
| Sequence-based reagent (qRT-PCR oligos) | RPL13A qRT-PCR oligo | This study | OMR069 | CAGGGTGGCTGTCACTGC |
| Sequence-based reagent (qRT-PCR oligos) | GAPDH q-RT-PCR | This study | OMR496 | CCTTTGACGCTGGGGCT |
| Sequence-based reagent (qRT-PCR oligos) | GAPDH q-RT-PCR | This study | OMR497 | GGTGGTCCAGGGGTCTT |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | OSBP2 | This study | OMR1589 | ccggaagcttACTGGCCGCTCGGCCGCGCGCGGGTCGGCCGGCTCTccaccATGacTTCGAAccgg |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | OSBP2 | This study | OMR1590 | ccggTTCGAAgtCATggtggAGAGCCGGCCGACCCGCGCGCGGCCGAGCGGCCAGTaagcttccgg |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | ctg uORF | This study | OMR1877 | ccggaCATACctgTatTCGATAATCAACTTTGAAAAACTCtaaa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | ctg uORF | This study | OMR1878 | CCGGtttaGAGTTTTTCAAAGTTGATTATCGAatAcagGTATGt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | gtg uORF | This study | OMR1879 | ccggaTGGAgtgAaaTCGATAATCAACTTTGAAAAACTCtaaa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | gtg uORF | This study | OMR1880 | CCGGtttaGAGTTTTTCAAAGTTGATTATCGAatAcacGTATGt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | ttg uORF | This study | OMR1881 | ccggaTGGAttgAaaTCGATAATCAACTTTGAAAAACTCtaaa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | ttg uORF | This study | OMR1882 | CCGGtttaGAGTTTTTCAAAGTTGATTATCGAatAcaaGTATGt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | TRAF7 | This study | OMR2182 | ccggaagcttGGCAGCCGTCCGGGC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | TRAF7 | This study | OMR2183 | ccggTTCGAAgtCATggtggGCTCTAGAGAGGCATCTACGGTCCTT |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | TEAD2 | This study | OMR2504 | AGCttCCCACTTTTCCCAAACAAAGCTCCCGGCAACTTTCTCCCTCGCAGCGCCCCGCCCGCCCGCGGCTCCCCAGCCCCAGGCCGGGAGGCCCAGcCATGACTT |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | TEAD2 | This study | OMR2505 | CGAAGTCATGgCTGGGCCTCCCGGCCTGGGGCTGGGGAGCCGCGGGCGGGCGGGGCGCTGCGAGGGAGAAAGTTGCCGGGAGCTTTGTTTGGGAAAAGTGGGa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | MXRA7 | This study | OMR2506 | agcttACTCGGCGGCC GCGGCGCGccatgactt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | MXRA7 | This study | OMR2507 | cgaagtcatggCGCGCCGCGGCCGCCGAGTa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CCDC125 | This study | OMR2508 | agcttGCGGCGGCAGCGGCGCACGCGCACGGAGAGGAGGCTACTTGCCAGACAGCCCATTTTTTCTTATGATAAAGACGGCATTTGGCTCccatgactt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CCDC125 | This study | OMR2509 | cgaagtcatggGAGCCAAATGCCGTCTTTATCATAAGAAAAAATGGGCTGTCTGGCAAGTAGCCTCCTCTCCGTGCGCGTGCGCCGCTGCCGCCGCa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | SCRN2 | This study | OMR2510 | agcttGCGGCCCTGGCCAGAAGCGGAGGAGGTGGCACCCGGGACCGAGCTGGGGTCTTGGAGGAAGAGAGGccatgactt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | SCRN2 | This study | OMR2511 | cgaagtcatggCCTCTCTTCCTCCAAGACCCCAGCTCGGTCC CGGGTGCCACCTCCTCCGCTTCTGGCCAGGGCCGCa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | RPS6KB2 | This study | OMR2512 | agcttAGTCAGTGCGCGGCCAGGTACGGGCCGACGGGCCCGCGGGGCCGGCGCCGCCccatgactt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | RPS6KB2 | This study | OMR2513 | cgaagtcatggGGCGGCGCCGGCCCCGCGGGCCCGTCGGCCCGTACCTGGCCGCGCACTGACTa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | SP140L | This study | OMR2514 | agcttACACTGCACGCAGGCTGGGCCGACTGGGGAGCTCATAGGCCAGGCTCTGACACCCAGGCAGGGCCTAGGGTGGGACGccatgactt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | SP140L | This study | OMR2515 | cgaagtcatggCGTCCCACCCTAGGCCCTGCCTGGGTGTCAGAGCCTGGCCTATGAGCTCCCCAGTCGGCCCAGCCTGCGTGCAGTGTa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CLPTM1L | This study | OMR2516 | agcttGACCCGGAGCGGGAAGccatgactt |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CLPTM1L | This study | OMR2517 | cgaagtcatggCTTCCCGCTCCGGGTCa |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | NCAPH2 | This study | OMR2518 | TAGTGAACCGTCAGATCACTAGAAGCTTGCATTTTCCTGGGCGGGAACAGCAAAATGGCGCCAGAACTAGTGGCGGGCTGAGGACGCCGTACCCCTCGGA |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | NCAPH2 | This study | OMR2519 | CTTTCGAAGTCATGGGTCCGGGAGGGAACGGGCGGCAAAGGGACCGCAGGGCTGCCTTCCGAGGGGTACGGCGTCCTCAGCCCGCCACTAGTTCTGGCGCC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CBX4 | This study | OMR2520 | AGAAGCTTAGTTGTCTGAGCGAGCGC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CBX4 | This study | OMR2521 | ACTTTCGAAGTCATGGGGCCGAGCCGGAGCG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | TEAD3 | This study | OMR2522 | AGAAGCTTAACACAAACTTTCCGTCCCGCTC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | TEAD3 | This study | OMR2523 | ACTTTCGAAGTCATGGTGTGCTGGTTGCTCTGGGC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | SLC7A11 | This study | OMR2526 | TAGAAGCttGGTTTGTAATGATAGGGCGGCAG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | SLC7A11 | This study | OMR2527 | ccggTTCGAAgtCATggtggAGTAGGGACACACGGGGG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | QTRT2 | This study | OMR2528 | TAGAAGCttAGTACTCCCTGATTGGCTCTGC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | QTRT2 | This study | OMR2529 | ccggTTCGAAgtCATggtggCCTAAGGGATTCTTCTAGGTCCTTTCAGC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | POLR3E | This study | OMR2530 | TAGAAGCttACGTGTCCGCCGGAGTT |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | POLR3E | This study | OMR2531 | ccggTTCGAAgtCATggtggACTAGAGGAGAGCCAGCCG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CNOT1 | This study | OMR2532 | TAGAAGCttGTAGAGAAACAAGCGGAGTTAACCGA |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CNOT1 | This study | OMR2533 | ccggTTCGAAgtCATggtggTGCTGGTTGGGGCGGAA |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | PNPLA8 | This study | OMR2536 | TAGAAGCttAGTGTTTGTGTTGGAAGCTCAGC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | PNPLA8 | This study | OMR2537 | ccggTTCGAAgtCATggtggAACTTAAAAATCATTTATTTTCTATGACATTCTCTCACTTCTTGA |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | ACTN4 | This study | OMR2538 | TAGAAGCttGAAGCAGCTGAAGCGGCG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | ACTN4 | This study | OMR2539 | ccggTTCGAAgtCATggtggTCCGCCGCCTCTCGC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | SLC38A1 | This study | OMR2540 | CTAgatatccaACTGACACGCAGCTTTGGTTAAA |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | SLC38A1 | This study | OMR2541 | ccggTTCGAAgtCATggtggGATTAGAAAGTGTCTGTAGTTTGAAAATTAGTCCA |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | short 5' UTR | This study | OMR2609 | CGTTTAGTGAACCGTCAGATCACCACCATGACTT |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | short 5' UTR | This study | OMR2610 | CGAAGTCATGGTGGTGATCTGACGGTTCACTAAACGAGCT |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | PIGG | This study | OMR277 | ccggaagcttGACGATAAGGCCTGGCG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | PIGG | This study | OMR278 | ccggTTCGAAgtCATggtggCGTGGACACGCTAGGCT |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | ATG9A | This study | OMR293 | ccggaagcttGAGTGGCAGACACCCG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | ATG9A | This study | OMR294 | ccggTTCGAAgtCATggtggCACCACCGCCCCCTG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | IQGAP1 | This study | OMR472 | ccggaagcttGACCCCGGCAAGCC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | IQGAP1 | This study | OMR473 | ccggTTCGAAgtCATggtggGGCGGACGAGCCC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | PTP4A1 | This study | OMR504 | ccggaagcttGAGATTACTGCCAGGCACA |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | PTP4A1 | This study | OMR505 | ccggTTCGAAgtCATggtggGTTAATTTAGTTAAAAAACACTCAATAGGGTTATGAA |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | IFRD1 | This study | OMR508 | ccggaagcttGTTAAAACCAGACTGCACTCC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | IFRD1 | This study | OMR509 | ccggTTCGAAgtCATggtggCGTGGGACGCCCGG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CCND3 | This study | OMR526 | ccggaagcttACCTATGCCGCGTGGG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | CCND3 | This study | OMR527 | CGAAGCGGCcgcATTTCACAATCATCTTTATTACAGTAGG |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | PPP1R15B | This study | OMR549 | ccggaagcttATTTTGGGCTTCGCTTCC |
| Sequence-based reagent (oligos used to clone 5'UTR of interest) | PPP1R15B | This study | OMR550 | ccggTTCGAAgtCATggtggACGGGATTCGGAGG |
| Sequence-based reagent (eIF2A Construct used for an overexpression) | N-terminally Flag-tagged eIF2A in pCDNA3 | This study | pMR007 | |
| Commercial assay or kit | Dual-Luciferase assay system | Promega | E1910 | |
| Commercial assay or kit | Cell Titer Glo | Promega | G7572 | |
| Commercial assay or kit | Next-Seq 550 system | Illumina | 20024906 | |
| Commercial assay or kit | Next-Flex small RNA v.4 kit protocol | Perkin Elmer | NOVA-5132–06 | |
| commercial assay or kit | Illumina TruSeq Stranded library preparation kit | Illumina | 20020594 | |
| Software, algorithm | algorithm to analyze ribosome profiling data | lab developed software Teleman, 2025 | https://github.com/aurelioteleman/Teleman-Lab | |
| Chemical compound, drug | sodium arsenite | Sigma | S7400-100G | |
| Chemical compound, drug | O-Propargyl-puromycin | Enzo Life Sciences | JBS-NU-931–05 | |
| Chemical compound, drug | tunicamycin | Sigma | 654380–10 MG |
Additional files
-
Supplementary file 1
Sequences of siRNA against eIF2A used in this study.
(Equimolar mix was used.).
- https://cdn.elifesciences.org/articles/105311/elife-105311-supp1-v1.xlsx
-
Supplementary file 2
Sequences of oligos used for sgRNA cloning.
- https://cdn.elifesciences.org/articles/105311/elife-105311-supp2-v1.xlsx
-
Supplementary file 3
Antibodies used in this study.
- https://cdn.elifesciences.org/articles/105311/elife-105311-supp3-v1.xlsx
-
Supplementary file 4
Sequences of oligos used for cloning in this study.
- https://cdn.elifesciences.org/articles/105311/elife-105311-supp4-v1.xlsx
-
Supplementary file 5
Transcript IDs from which 5'UTRs were cloned in this study.
- https://cdn.elifesciences.org/articles/105311/elife-105311-supp5-v1.xlsx
-
Supplementary file 6
Sequences of Q-RT-PCR oligos used in this study.
- https://cdn.elifesciences.org/articles/105311/elife-105311-supp6-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/105311/elife-105311-mdarchecklist1-v1.docx
-
Source data 1
Source data underlying all graphs in the paper.
- https://cdn.elifesciences.org/articles/105311/elife-105311-data1-v1.xlsx





