The myeloid cell-driven transdifferentiation of endothelial cells into pericytes promotes the restoration of BBB function and brain self-repair after stroke
Figures
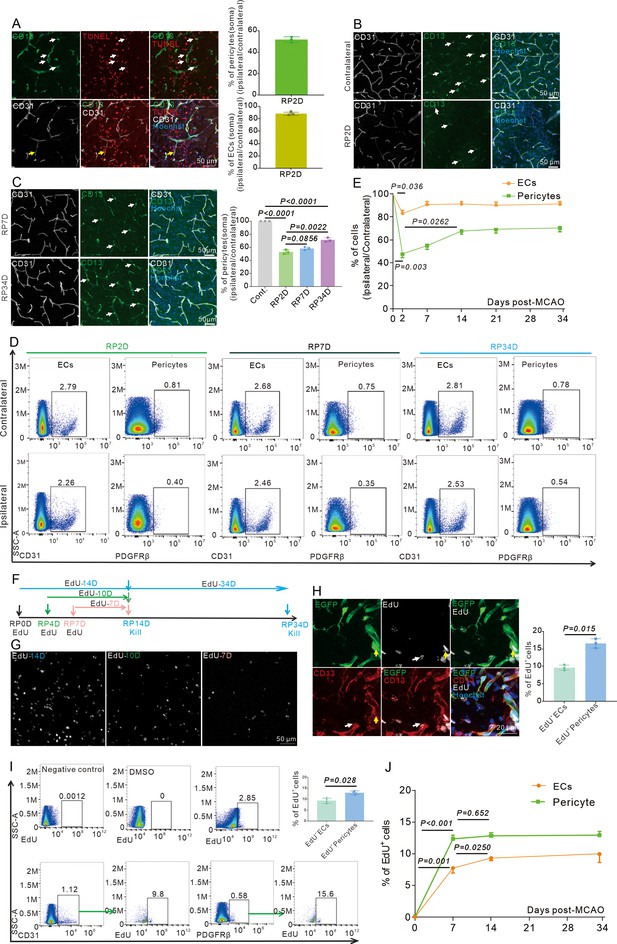
Pericytes die rapidly in the acute phase and replenish in the subacute and chronic phases after stroke.
(A) Immunoflurescence staining shows Tunel+ pericytes (white) and Tunel+ endothelial cells (ECs) (yellow) after middle cerebral artery occlusion (MCAO) at RP2D and quantifies the proportion of Tunel+ cells in pericytes and ECs (n = 3, 20 slices/mouse). (B) Immunofluorescence staining shows CD13+ soma after MCAO at RP2D (n = 3, 20 slices/mouse). (C) Immunofluorescence staining shows CD13+ soma after MCAO at RP7D and RP34D, quantifying the ratio of CD13+ soma (n = 3, 20 slices/mouse). (D) Flow cytometry analysis of the proportion of pericytes and ECs after MCAO at RP2D, RP7D, and RP34D (n = 6). (E) Quantitative analysis of the proportion of pericytes and ECs at different reperfusion times after stroke (n = 6). (F) Schematic diagram displaying the time course for EdU injection and analysis time points. (G) Maximum EdU signal in the ischemic area at different EdU injection times (n = 3). (H) Immunofluorescence staining shows EdU+ pericytes (white) and EdU+ ECs (yellow) after MCAO at RP34D and quantifies the proportion of EdU+ cells in pericytes and ECs (n = 3, 20 slices/mouse). (I) Flow cytometry analysis of the proportion of EdU+ pericytes and EdU+ ECs after MCAO at RP14D (n = 4). (J) Flow cytometry analysis of the proportion of EdU+ pericytes and EdU+ ECs after MCAO at RP7D, RP14, and RP34D (n = 4). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (C, E, H and J).
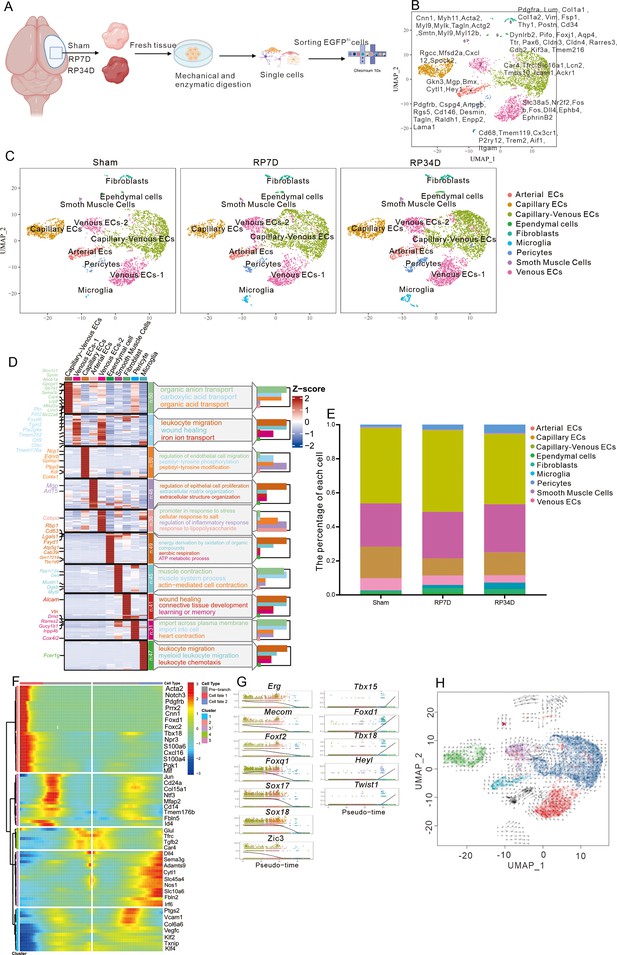
scRNA-seq is used to explore the fate of endothelial cells (ECs) after stroke.
(A) Samples were obtained from mouse ischemic brains at sham, RP7D, and RP34D. Single cells were processed using Chromium 10x 3′DEG chemistry. (B) Uniform manifold approximation and projection (UMAP) embedding of all cells and marker genes. (C) UMAP analysis of individual sham, RP7D, and RP34D cell transcriptomes showed 10 clusters. (D) Heatmap displays the expression of the top 50 upregulated genes in each cluster. The scale bar represents the z-score of average gene expression (log). (E) Relative proportion of major cell types in different reperfusion times. (F) Differential gene expression variance over pseudotime of ECs differentiation trajectory branches. (G) Differential transcription factors expression variance over pseudotime of ECs differentiation trajectory branches. (H) Pseudotime trajectory of ECs differentiation trajectory branches. The arrows show the direction of pseudotime trajectories.
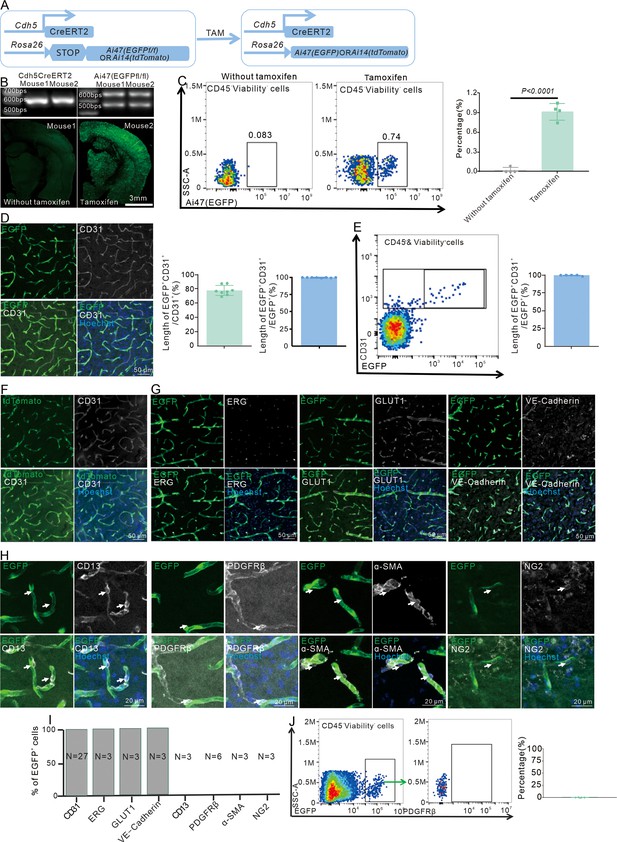
Cdh5CreERT2 mice specifically recombine parenchymal endothelial cells (ECs).
(A) Schematic diagram displaying Cdh5CreERT2 mice label ECs. (B) Immunofluorescence staining shows with or without tamoxifen in Cdh5Cre-ERT2;Ai47 mice (n = 3, 10 slices/mouse). (C) Flow cytometry analyzes the proportion of EGFP+ cells and quantifies the proportion of EGFP+ cells (n = 4). (D) Immunofluorescence staining of CD31 expression in Cdh5CreERT2;Ai47 mice and quantitative analysis of the proportion of EGFP+&CD31+/CD31+ or EGFP+ blood vessel (n = 8, 10 slices/mouse). (E) Flow cytometry analysis of the proportion of EGFP+&CD31+/EGFP+ cells and quantitative analysis of the proportion (n = 5). (F) Immunofluorescence staining of CD31 expression in Cdh5CreERT2;Ai14 mice (n = 3, 20 slices/mouse). (G) Immunofluorescence staining of ECs markers (ERG, GLUT1, and VE-Cadherin) expression in Cdh5CreERT2;Ai47 mice (n = 3–27, 10 slices/mouse). (H) Immunofluorescence staining of pericyte markers (CD13, PDGFRβ, α-SMA, and NG2) expression in Cdh5CreERT2; Ai47 mice (n = 3–6, 20 slices/mouse). (I) Quantitative analysis of the proportion of F, G, and H. (J) Flow cytometry analysis of the proportion of PDGFRβ+ cells in EGFP+ cells and quantitative analysis of the proportion (n = 4). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (C).
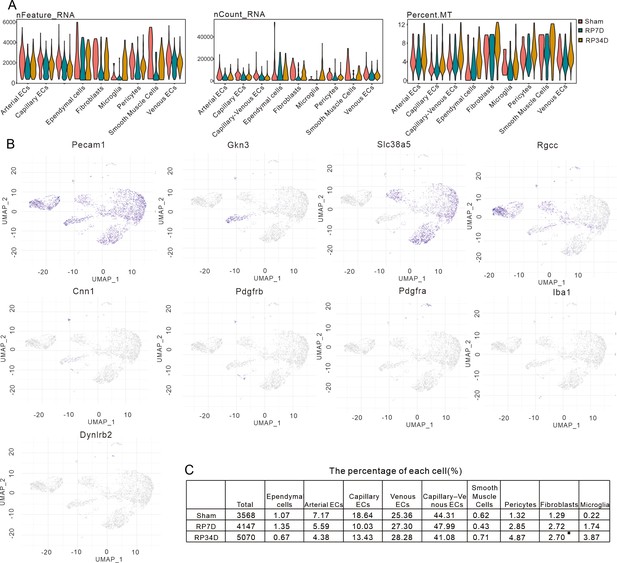
EGFP+ cells transcriptomic dataset quality.
(A) Violin plots showing the distribution of the number of total UMI counts per cell (nCount), genes detected per cell (nFeature), and percentage of mitochondrial genes (percent.mt) per identified cell type. (B) Uniform manifold approximation and projection (UMAP) plots depicting the expression of individual marker genes for endothelial cells (ECs) (Pecam1), arterial ECs (Gkn3), venous ECs (Slc38a5), capillary ECs (Rgcc), smooth muscle cells (SMCs) (Cnn1), pericytes (Pdgfrb), fibroblasts (Pdgfra), microglia (Iba1), and ependymal cells (Dynlrb2). Scale bars represent the log of normalized gene expression. (C) Statistical table showing the percentage and number for each cell type.
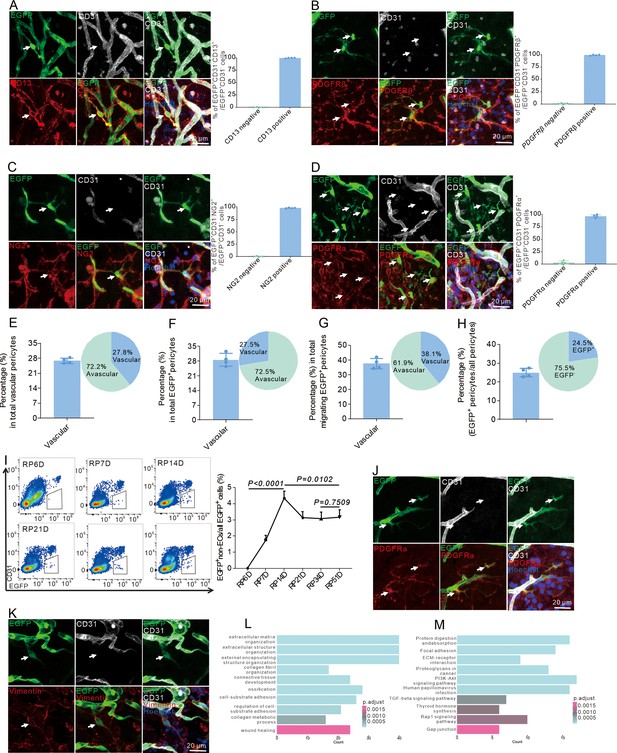
Endothelial-to-pericytic transition can replenish pericytes and undergo an intermediate fibroblast-like cell state after stroke.
(A) Immunofluorescence staining of CD31 and CD13 expression in Cdh5CreERT2;Ai47 mice with middle cerebral artery occlusion (MCAO) at RP34D and quantitative analysis of the proportion of CD13+ cells (n = 5, 20 slices/mouse). (B) Immunofluorescence staining of CD31 and PDGFRβ expression in Cdh5CreERT2;Ai47 mice with MCAO at RP34D and quantitative analysis of the proportion of PDGFRβ+ cells (n = 5, 20 slices/mouse). (C) Immunofluorescence staining of CD31 and NG2 expression in Cdh5CreERT2;Ai47 mice with MCAO at RP34D and quantitative analysis of the proportion of NG2+ cells (n = 5, 20 slices/mouse). (D) Immunofluorescence staining of CD31 and PDGFRα expression in Cdh5CreERT2;Ai47 mice with MCAO at RP34D and quantitative analysis of the proportion of PDGFRα+ cells (n = 5, 20 slices/mouse). (E) Quantitative analysis of the proportion of EGFP+ pericytes on blood vessel/pericytes on blood vessel (n = 5, 20 slices/mouse). (F) Quantitative analysis of the proportion of EGFP+ pericytes on blood vessel/EGFP+ pericytes (n = 5, 20 slices/mouse). (G) Quantitative analysis of the proportion of EGFP+ pericytes on blood vessels/migrating EGFP+ pericytes (n = 5, 20 slices/mouse). (H) Quantitative analysis of the proportion of EGFP+ pericytes/all pericytes (n = 5, 20 slices/mouse). (I) Flow cytometry analysis of the proportion of EGFP+&CD31- cells after MCAO at different times and quantitative analysis of the proportion of EGFP+&CD31- cells (n = 4). (J) Immunofluorescence staining of CD31 and PDGFRα expression in Cdh5CreERT2;Ai47 mice with MCAO at RP8D (n = 5, 20 slices/mouse). (K) Immunofluorescence staining of CD31 and vimentin expression in Cdh5CreERT2;Ai47 mice with MCAO at RP8D (n = 5, 20 slices/mouse). (L) Gene Ontology (GO) analysis of upregulated differentially expressed genes (DEGs) in EGFP+&CD31- cells subgroup, compared with EGFP+&CD31+ cells from contralateral. (M) KEGG pathway analysis of upregulated DEGs in EGFP+&CD31- cells subgroup, compared with EGFP+&CD31+ cells from contralateral. Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (I).
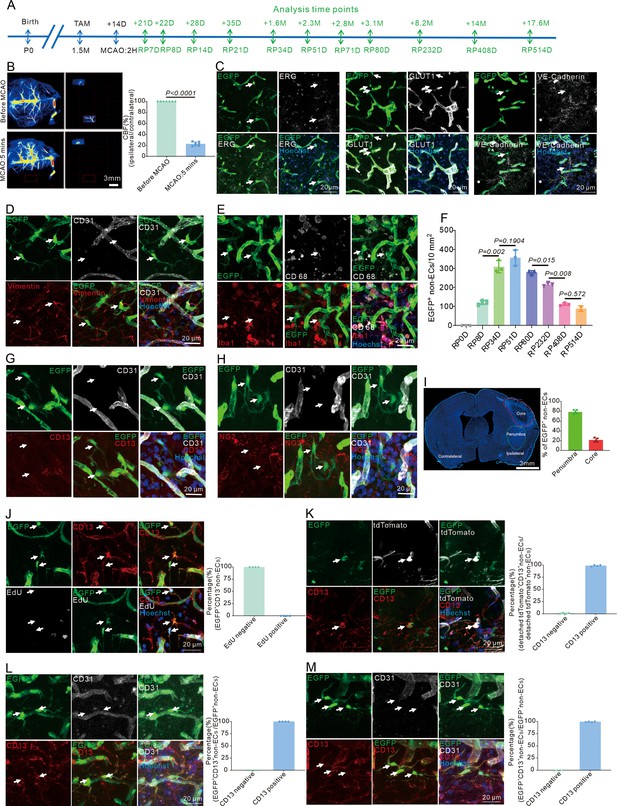
Different means prove that endothelial cells (ECs) can give rise to pericyte-like cells after stroke.
(A) Schematic diagram displaying the time course for middle cerebral artery occlusion (MCAO), tamoxifen, and analysis time points. (B) Laser speckle displaying cerebral blood flow (CBF) change after stroke (ipsilateral/contralateral) and quantifying the proportion (n = 7). (C) Immunofluorescence staining of ECs markers (ERG, GLUT1, and VE-Cadherin) expression in Cdh5CreERT2;Ai47 mice with MCAO after RP34D (n = 3, 20 slices/mouse). (D) Immunofluorescence staining of CD31 and vimentin expression in Cdh5CreERT2;Ai47 mice with MCAO at RP34D. (E) Immunofluorescence staining of CD68 and Iba1 expression in Cdh5CreERT2;Ai47 mice with MCAO at RP34D. (F) Quantitative analysis of the number of EGFP+&CD31- cells in unit area at different reperfusion times after stroke (n = 2–6). (G) Immunofluorescence staining of CD31 and CD13 expression in Cdh5CreERT2;Ai47 mice with MCAO at RP8D. (H) Immunofluorescence staining of CD31 and NG2 expression in Cdh5CreERT2;Ai47 mice with MCAO at RP8D. (I) Quantitative analysis of the percentage of EGFP+&CD31- cells in core and penumbra after stroke at RP34D (n = 4, 20 slices/mouse). (J) Immunofluorescence staining of EdU expression in EGFP+ pericytes after MCAO at RP34D and quantitative analysis of the proportion of EdU+ cells (n = 4, 20 slices/mouse). (K) Immunofluorescence staining of CD31 and CD13 expression in Cdh5CreERT2 mice injected with AAV2/9-CAG-DIO-EGFP virus after MCAO at RP34D and quantitative analysis of the proportion of CD13+ cells (n = 4, 20 slices/mouse). (L) Immunofluorescence staining of CD31 and CD13 expression in Tie2Dre;Mfsd2aXER;Ai47 mice with MCAO at RP34D and quantitative analysis of the proportion of CD13+ cells (n = 4, 20 slices/mouse). (M) Immunofluorescence staining of CD31 and CD13 expression in Ai47 mice infected with AAV-BI30-Cre virus after MCAO at RP34D and quantitative analysis of the proportion of CD13+ cells (n = 3, 20 slices/mouse). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (B and F).
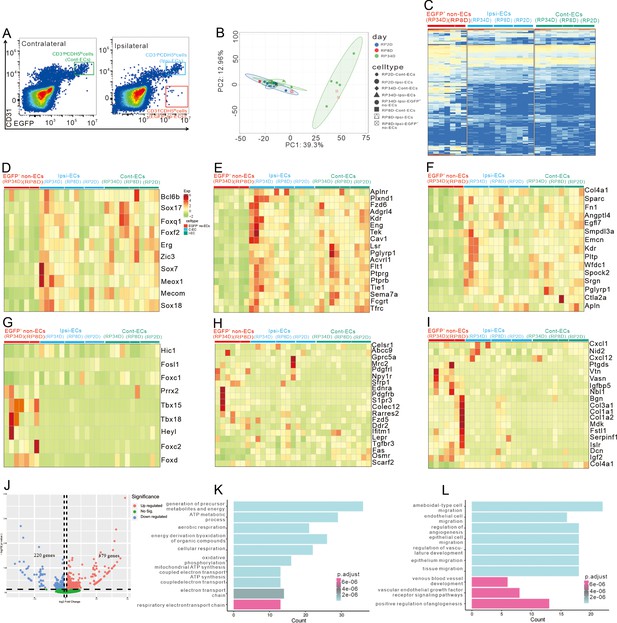
Endothelial cells (ECs) give rise to cells with a similar pericyte transcriptome profile after stroke.
(A) The gate of sorting ECs in contralateral (C-ECs), ECs in ipsilateral (I-ECs), and EGFP+&CD31- cells in ipsilateral (EGFP+ non-ECs). (B) Principal component analysis (PCA) of the variance-stabilized estimated raw counts of differentially expressed genes (DEGs). (C) The heatmap showing 100 genes in all groups. (D) The heatmap showing EC transcription factor genes expression in all groups (n = 2–5). (E) The heatmap showing endothelial-enriched transmembrane receptor genes expression in all groups (n = 2–5). (F) The heatmap showing endothelial-enriched ligand genes expression in all groups (n = 2–5). (G) The heatmap showing pericytic transcription factors genes expression in all groups (n = 2–5). (H) The heatmap showing pericytic-enriched transmembrane receptor gene expression in all groups (n = 2–5). (I) The heatmap showing pericytic-enriched ligand genes expression in all groups (n = 2–5). (J) Volcano plot showing differential expression of genes in EGFP+ non-ECs/C-ECs (n = 5). (K) Gene Ontology (GO) functional enrichment analysis from upregulated genes in DEG. (L) KEGG functional enrichment analysis from upregulated genes in DEG.
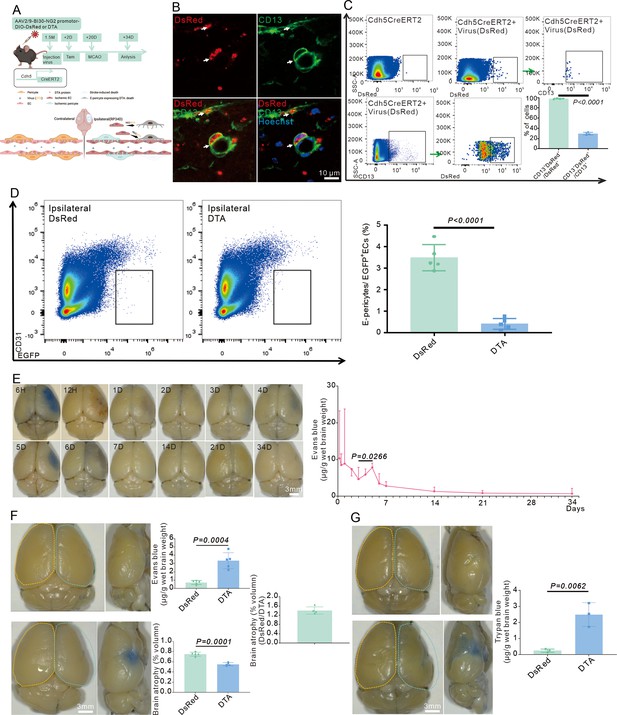
E-pericytes deletion by AAV2/9-BI30-DIO-NG2-promotor-DTA virus aggravates blood-brain barrier (BBB) leakage after stroke.
(A) Schematic diagram displaying Cdh5CreERT2 injected with AAV2/9-BI30-DIO-NG2-promotor -DsRed or DTA virus to kill the cell and the time course for tamoxifen, middle cerebral artery occlusion (MCAO), and analysis time points. (B) Immunofluorescence staining of CD13 expression in Cdh5CreERT2 injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed A virus (n = 3). (C) Flow cytometry analysis of the proportion of DsRed+&CD13+/DsRed+ cells and quantitative analysis of the proportion (n = 3). (D) Flow cytometry analysis of the proportion of E-pericytes cells in Cdh5CreERT2 injected with AAV2/9-BI30-DIO-NG2-promotor-DTA virus and quantitative analysis of the proportion (n = 5). (E) Image showing the leakage of Evans blue in wild-type (WT) mice at different times after MCAO and quantitative analysis of the leakage of Evans blue (n = 3). (F) Image showing the leakage of Evans blue in Cdh5CreERT2 injected with AAV2/9-BI30-DIO-NG2-promotor-DTA virus at RP34D after MCAO, quantitative analysis of the leakage of Evans blue (n = 5), and the brain atrophy volume (n = 5). (G) Image showing the leakage of trypan blue in Cdh5CreERT2 injected with AAV2/9-BI30-DIO-NG2-promotor-DTA virus at RP34D after MCAO and quantitative analysis of the leakage of trypan blue (n = 3). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (C, D, F and G).
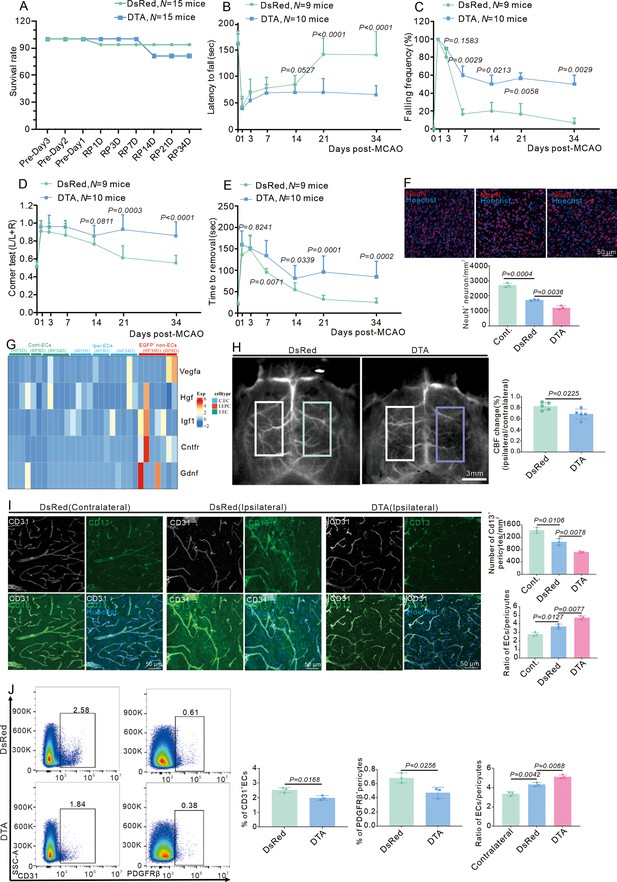
E-pericytes deletion by AAV2/9-BI30-DIO-NG2-promotor-DTA virus exacerbates neurological deficit after stroke.
(A) Graph showing the survival rate of mice in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed or DTA virus after middle cerebral artery occlusion (MCAO) at RP34D (n = 15). (B) Graph showing rotarod test in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed or DTA virus after MCAO (n = 9–10). (C) Graph showing beam walking test in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed or DTA virus after MCAO (n = 9–10). (D) Graph showing corner test in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed or DTA virus after MCAO (n = 9–10). (E) Graph showing adhesive movement test in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed or DTA virus after MCAO (n = 9–10). (F) Immunofluorescence staining of NeuN expression in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-Long-DsRed or DTA virus after MCAO at RP34D and quantitative analysis of the number in the unit area (n = 4, 20 slices/mouse). (G) The heatmap showing promotion neuron survival and growth genes expression in all groups (n = 5). (H) Image showing the change of cerebral blood flow (CBF) in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed or DTA virus after MCAO at RP34D and quantitative analysis of the change of CBF (n = 5). (I) Immunofluorescence staining of CD13 and CD31 expression in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed or DTA virus after MCAO at RP34D and quantitative analysis of the percentage (n = 3, 20 slices/mouse). (J) Flow cytometry analysis of the proportion of CD13+ cells and CD31+ cells in Cdh5CreERT2 mice injected with AAV2/9-BI30-DIO-NG2-promotor-DsRed or DTA virus after MCAO at RP34D and quantitative analysis of the percentage (n = Immunofluorescence staining of p-SMAD3 expression 3). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A-F and H-J).
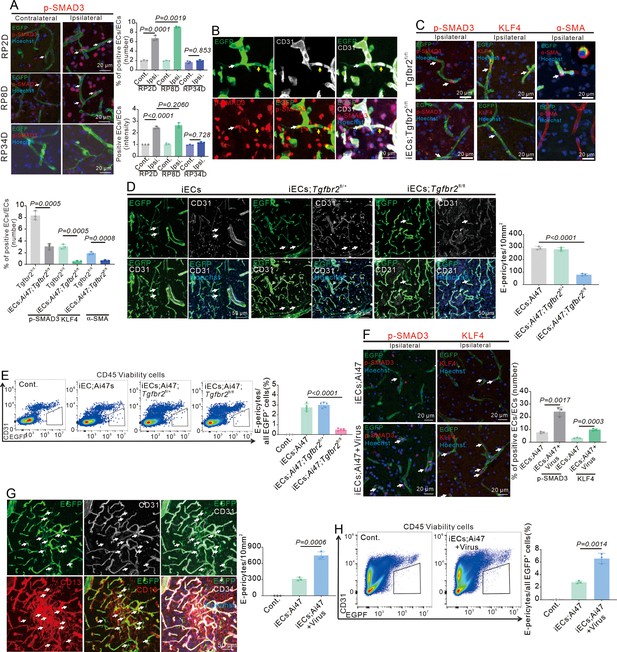
Endothelial-specific loss and reinforcement of Tgfbr2 gene expression affect endothelial-mesenchymal transformation (EndoMT) and E-pericytes.
(A) Immunofluorescence staining of p-SMAD3 expression in Cdh5CreERT2;Ai47 mice at RP2D, RP8D, and RP34D and quantitative analysis of the proportion and intensity (n = 3, 20 slices/mouse). (B) Immunofluorescence staining of CD31 and p-SMAD3 expression in Cdh5CreERT2;Ai47 mice at RP8D (white:EGFP+&p-SMAD3+&CD31-; yellow:EGFP+&p-SMAD3-&CD31+; n = 3). (C) Immunofluorescence staining of EndoMT markers (p-SMAD3, KLF4, and α-SMA) expression in Cdh5CreERT2;Ai47;Tgfbr2fl/fl mice at RP2D and quantitative analysis of the proportion and intensity (n = 3, 20 slices/mouse). (D) Immunofluorescence staining of CD31 expression in Cdh5CreERT2;Ai47;Tgfbr2fl/fl mice at RP34D and quantitative analysis of the number in the unit area (n = 3, 20 slices/mouse). (E) Flow cytometry analysis of the proportion of E-pericytes in Cdh5CreERT2;Ai47;Tgfbr2fl/fl mice at RP34D and quantitative analysis of the proportion (n = 5). (F) Immunofluorescence staining of EndoMT markers (p-SMAD3+ and KLF4+) expression in Cdh5CreERT2;Ai47 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs at RP2D and quantitative analysis of the proportion of EndoMT markers (n = 3, 20 slices/mouse). (G) Immunofluorescence staining of CD31 and CD13 expression in Cdh5CreERT2;Ai47 mice with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs at RP34D and quantitative analysis of the number in unit area (n = 3, 20 slices/mouse). (H) Flow cytometry analysis of the proportion of E-pericytes in Cdh5Cre ERT2;Ai47 mice with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs at RP34D and quantitative analysis of the proportion of E-pericytes (n = 3). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A and C-H).
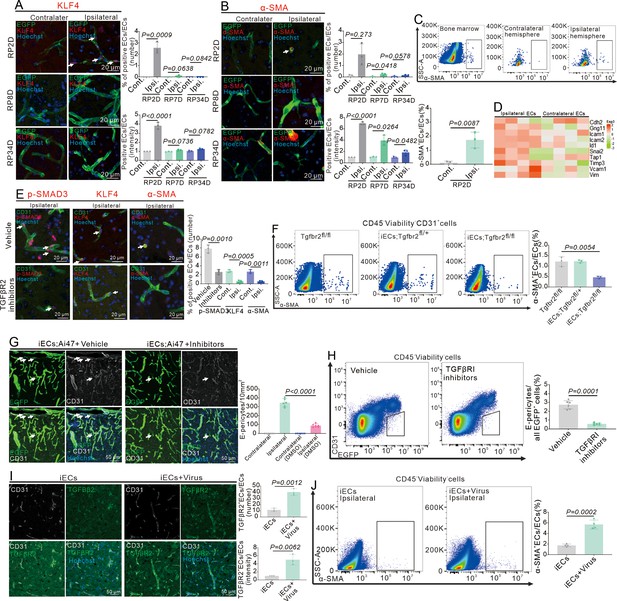
Systemic inhibition of TGFβR2 reduces endothelial-mesenchymal transformation (EndoMT) and E-pericytes.
(A) Immunofluorescence staining of KLF4 expression in Cdh5CreERT2;Ai47 mice at RP2D, RP8D, and RP34D and quantitative analysis of the proportion and intensity (n = 3, 20 slices/mouse). (B) Immunofluorescence staining of ɑ-SMA expression in Cdh5CreERT2;Ai47 mice at RP2D, RP8D, and RP34D and quantitative analysis of the proportion and intensity (n = 3, 20 slices/mouse). (C) Flow cytometry analysis of the proportion of ɑ-SMA+&CD31+ endothelial cells (ECs) in Cdh5CreERT2;Ai47 mice at RP2D and quantitative analysis of the proportion (n = 3). (D) Heatmap depiction of different EndoMT marker genes in ECs at RP2D (n = 4). (E) Immunofluorescence staining of EndoMT markers (p-SMAD3, KLF4, and α-SMA) expression in Cdh5CreERT2;Ai47 mice injected with TGFβR2 inhibitors at RP2D and quantitative analysis of the proportion (n = 3, 20 slices/mouse). (F) Flow cytometry analysis of the proportion of ɑ-SMA+&CD31+ ECs in Cdh5CreERT2;Ai47;Tgfbr2fl/fl mice at RP2D and quantitative analysis of the proportion (n = 3). (G) Immunofluorescence staining of CD31 expression in Cdh5CreERT2;Ai47 mice injected with TGFβR2 inhibitors at RP34D and quantitative analysis of the number in the unit area (n = 3, 20 slices/mouse). (H) Flow cytometry analysis of the proportion of E-pericytes in Cdh5CreERT2;Ai47 mice injected with TGFβR2 inhibitors at RP34D and quantitative analysis of the proportion (n = 3). (I) Immunofluorescence staining of CD31 and TGFβR2 expression in Cdh5CreERT2;Ai47 mice injected with AAV2/9-BI30-EF1α-DIO- Tgfbr2-3XFLAG-P2A-DsRed-WPREs and quantitative analysis of the proportion and intensity (n = 3,20 slices/mouse). (J) Flow cytometry analysis of the proportion of ɑ-SMA+&CD31+ ECs in Cdh5CreERT2;Ai47 mice with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs at RP2D and quantitative analysis of the proportion. Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A-C and E-J).
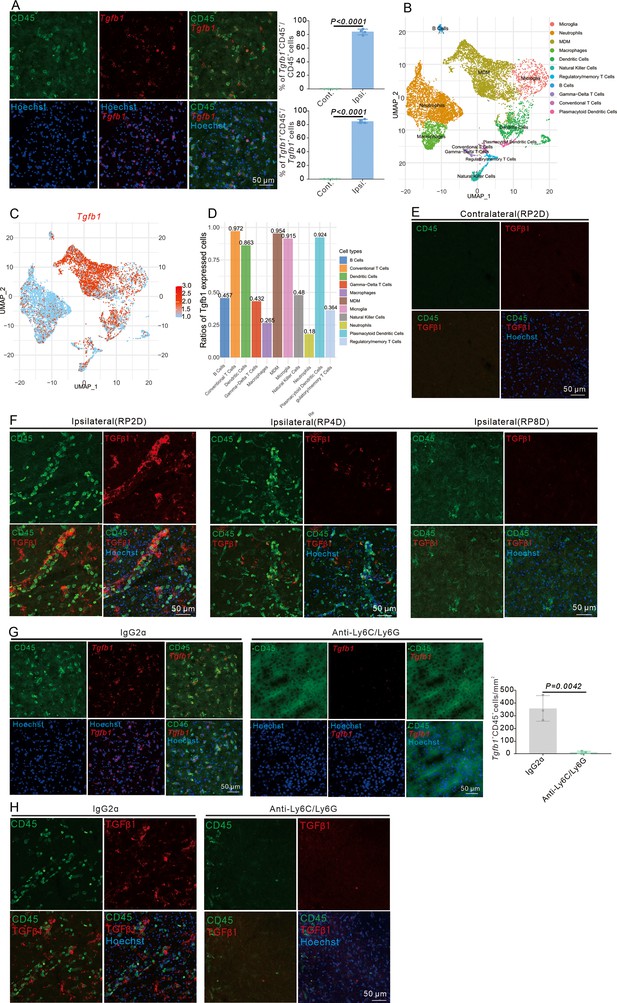
Infiltrating myeloid cells produce dominant TGFβ1 in mouse brain after stroke.
(A) Immunofluorescence staining of CD45 and Tgfb1 gene expression in wild-type (WT) mice with middle cerebral artery occlusion (MCAO) at RP2D and quantitative analysis of the proportion of Tgfb1+ cells (n = 4, 10 slices/mouse). (B) scRNA-seq of CD45+ cells isolated from the ipsilateral brain with MCAO:2H and RP1.5D. Dimensionality reduction and identification of clusters of transcriptionally similar cells were performed in an unsupervised manner (n = 1), monocyte-derived macrophage (MDM). (C) Tgfb1 gene expression in dimensionality reduction and identification of clusters (n = 1). (D) The percentage of Tgfb1 gene expression in different clusters (n = 1). (E) TGFβ1 protein expression in contralateral brain with MCAO at RP2D (n=3,20 slices/mouse). (F) TGFβ1 protein expression in the ipsilateral brain with MCAO at RP2D, RP4D, and RP8D (n = 3, 20 slices/mouse). (G) Immunofluorescence staining of CD45 and Tgfb1 gene expression in WT mice injected with anti-Ly6C/Ly6G after MCAO at RP2D and quantitative analysis of the proportion of CD45+&Tgfb1+ cells in the unit area (n = 4,10 slices/mouse). (H) Immunofluorescence staining of CD45 and TGFβ1 protein expression in WT mice injected with anti-Ly6C/Ly6G after MCAO at RP2D. Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A and G).
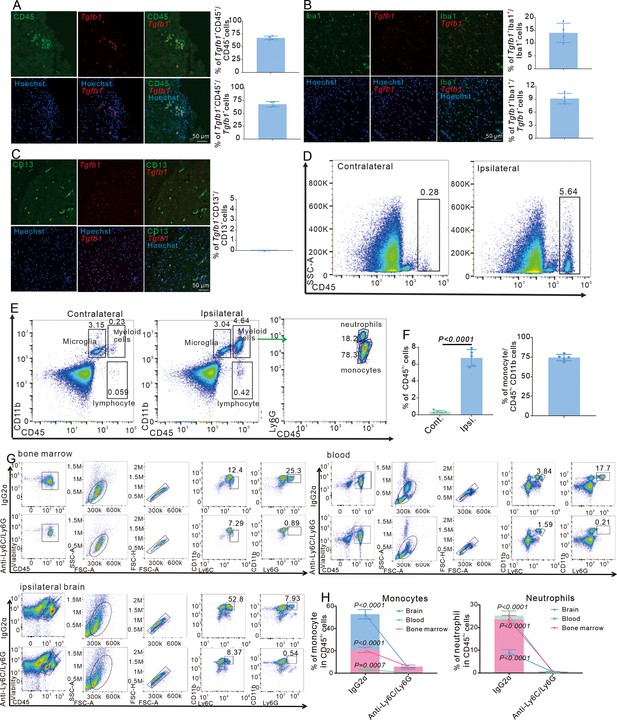
Myeloid cells are leading immune cells in the mouse brain after stroke at RP2D.
(A) Immunofluorescence staining of CD45 and Tgfb1 gene expression in wild-type (WT) mice with middle cerebral artery occlusion (MCAO) at RP1D and quantitative analysis of the proportion of Tgfb1+ cells (n = 4, 10 slices/mouse). (B) Immunofluorescence staining of Iba1 and Tgfb1 gene expression in WT mice with MCAO at RP2D and quantitative analysis of the proportion of Tgfb1+ cells (n = 4, 10 slices/mouse). (C) Immunofluorescence staining of CD13 and Tgfb1 gene expression in WT mice with MCAO at RP2D and quantitative analysis of the proportion of Tgfb1+ cells (n = 4, 10 slices/mouse). (D) Flow cytometry analysis of the proportion of CD45hi cells from ipsilateral brain with MCAO:2H and RP1.5D. (E) Flow cytometry analysis of the proportion of myeloid cells from the ipsilateral brain with MCAO:2H and RP1.5D. (F) Quantitative analysis of the proportion of CD45hi cells (D) and monocyte (E) (n = 5). (G) Flow cytometry analysis of the proportion of myeloid cells from bone marrow, blood, and ipsilateral brain after mice injected with anti-Ly6C/Ly6G at RP2D (n = 4). (H) Quantitative analysis of the proportion of monocyte and neutrophil in (J) (n = 4). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A, B, F and H).
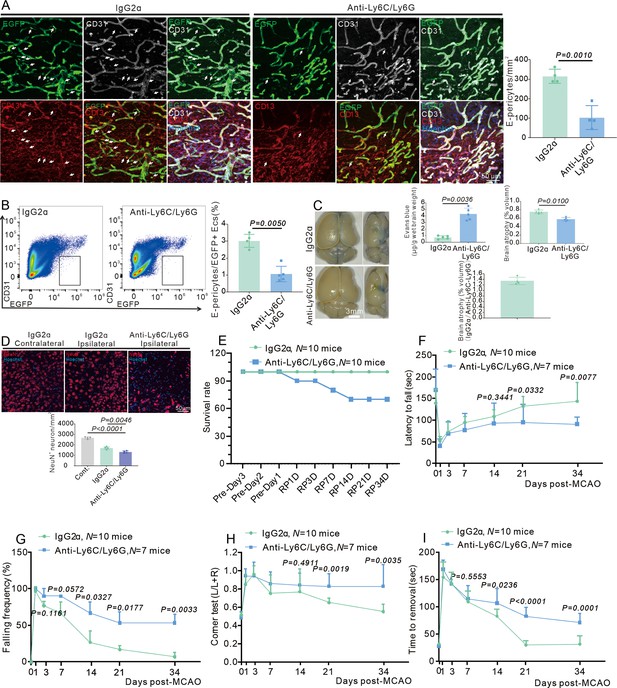
Infiltrating myeloid cells promote blood-brain barrier (BBB) renovation and neurological recovery after stroke.
(A) Immunofluorescence staining of CD13 and CD31 expression in wild-type (WT) mice injected with anti-Ly6C/Ly6G at RP34D after middle cerebral artery occlusion (MCAO) and quantitative analysis of the number in the unit area (n = 4, 20 slices/mouse). (B) Flow cytometry analysis of the proportion of EGFP+&CD31+ cells in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO and quantitative analysis of the percentage (n = 4). (C) Image showing the leakage of Evans blue in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO and quantitative analysis of the leakage of Evans blue and brain atrophy volume (n = 5). (D) Immunofluorescence staining of NeuN expression in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO and quantitative analysis of the number in the unit area (n = 4, 20 slices/mouse). (E) Graph showing the survival rate of mice in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO (n = 10). (F) Graph showing rotarod test in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO (n = 7–10). (G) Graph showing beam walking test in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO (n = 7–10). (H) Graph showing corner test in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO (n = 7–10). (I) Graph showing adhesive movement test in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO (n = 7–10). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A-D and F-I).
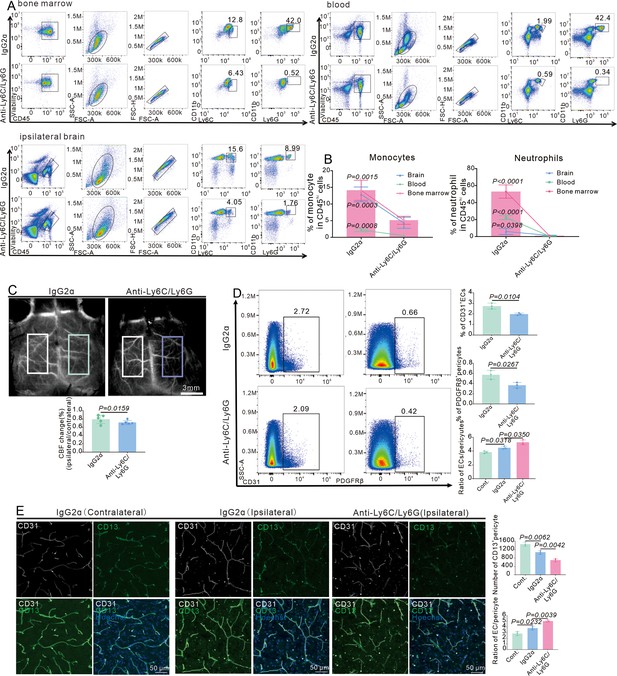
Infiltrating myeloid cells promote vascular function recovery after stroke.
(A) Flow cytometry analysis of the proportion of myeloid cells from bone marrow, blood, and ipsilateral brain after mice injected with anti-Ly6C/Ly6G at RP34D (n = 4). (B) Quantitative analysis of the proportion of monocyte and neutrophil in (A) (n = 4). (C) Image showing the change of cerebral blood flow (CBF) in wild-type (WT) mice injected with anti-Ly6C/Ly6G at RP34D after middle cerebral artery occlusion (MCAO) and quantitative analysis of the change of CBF (n = 5). (D) Flow cytometry analysis of the proportion of CD13+ cells and CD31+ cells in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO and quantitative analysis of the percentage (n = 3). (E) Immunofluorescence analysis of the proportion of CD13+ cells and CD31+ cells in WT mice injected with anti-Ly6C/Ly6G at RP34D after MCAO and quantitative analysis of the percentage (n = 3). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (B-E).
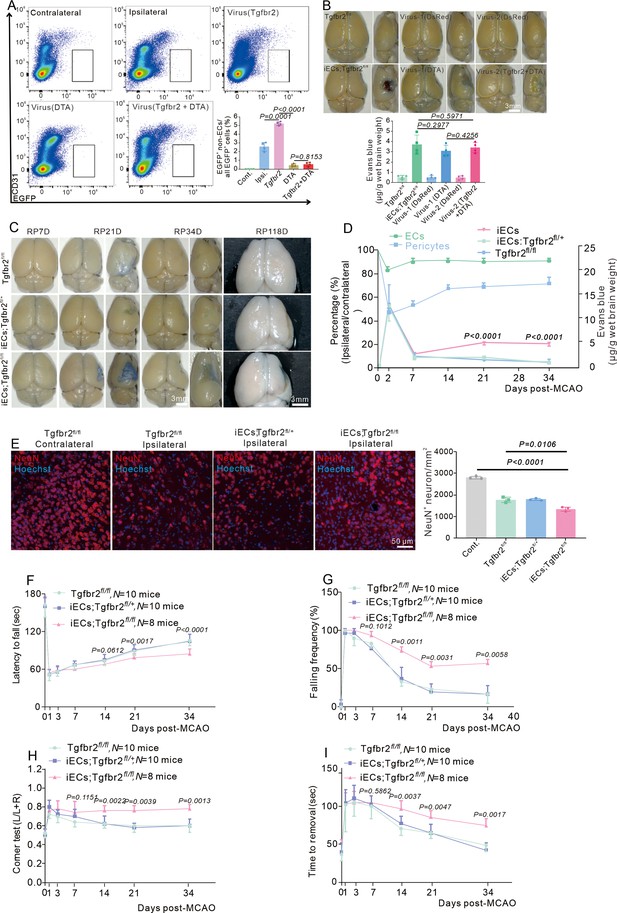
E-pericytes deletion by specific endothelial cells (ECs) knockout of the Tgfbr2 gene aggravates blood-brain barrier (BBB) leakage and neurological deficit after stroke.
(A) Flow cytometry analysis of the proportion of E-pericytes synchronous injection with AAV2/9-BI30-DIO-NG2-promotor diphtheria toxin A (DTA) virus and AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after middle cerebral artery occlusion (MCAO) at 14D (n = 4). (B) Image showing the leakage of Evans blue in induced ECs (iECs);Tgfbr2fl/fl mice at different times after MCAO and quantitative analysis of the leakage of Evans blue (n = 3). (C) Graph showing the leakage of Evans blue in iECs;Tgfbr2fl/fl mice at different times after MCAO and quantitative analysis of the leakage of trypan blue (n = 3). (D) Graph showing the percentage of pericyte and ECs, the leakage of Evans blue in iECs;Tgfbr2fl/fl mice at different times after MCAO (n = 3–6). (E) Immunofluorescence staining of NeuN expression in iECs;Tgfbr2fl/fl mice at RP34D after MCAO and quantitative analysis of the number of neurons in the unit area (n = 3, 20 slices/mouse). (F) Graph showing the rotarod test in iECs;Tgfbr2fl/fl mice after MCAO at RP34D (n = 8–10). (G) Graph showing the beam walking test in iECs;Tgfbr2fl/fl mice after MCAO at RP34D (n = 8–10). (H) Graph showing the corner test in iECs;Tgfbr2fl/fl mice after MCAO at RP34D (n = 8–10). (I) Graph showing the adhesive movement test in iECs;Tgfbr2fl/fl mice after MCAO at RP34D (n = 8–10). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A-B and D-I).
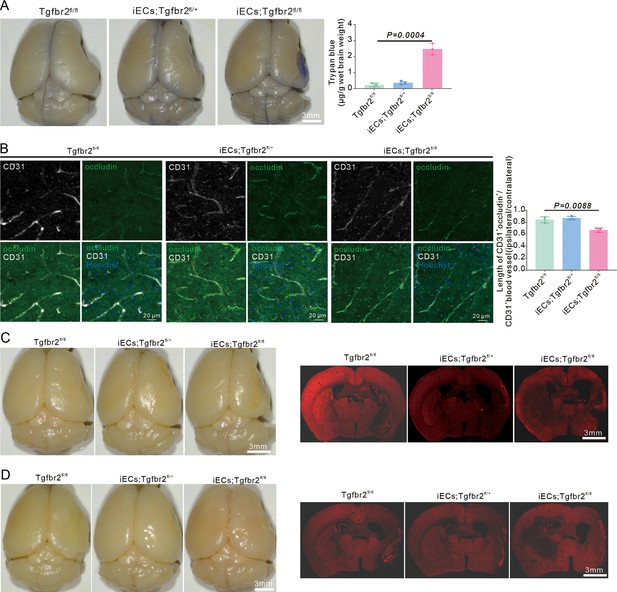
E-pericytes deletion by specific endothelial cells (ECs) knockout of the Tgfbr2 gene aggravates blood-brain barrier (BBB) leakage of small molecules.
(A) Image showing the leakage of trypan blue in induced ECs (iECs);Tgfbr2fl/fl mice at RP34D after middle cerebral artery occlusion (MCAO) and quantitative analysis of the leakage of trypan blue (n = 3). (B) Image showing the leakage of dextran-rhodamine B in iECs;Tgfbr2fl/fl mice at RP34D after MCAO (n = 3, 10 slices/mouse). (C) Image showing the leakage of Texas-Ted in iECs;Tgfbr2fl/fl mice at RP34D after MCAO (n = 3, 20 slices/mouse). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A and B).
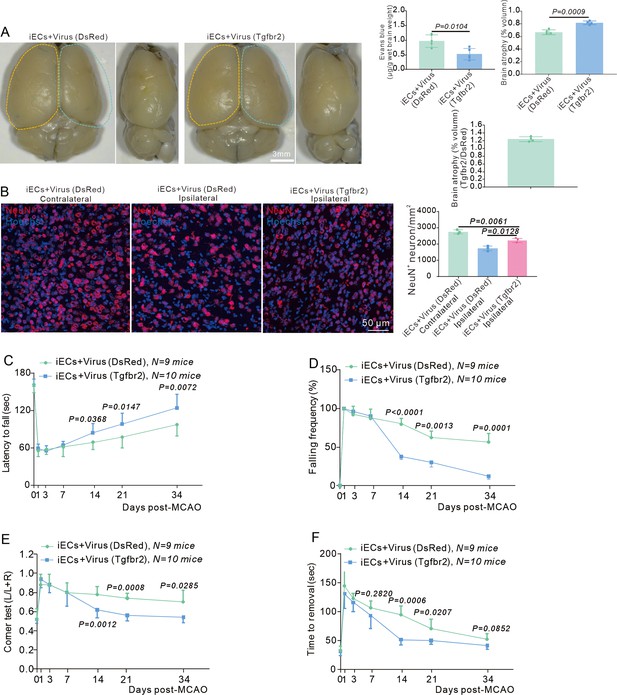
Endothelial cell (EC)-specific overexpression of the Tgfbr2 gene reinforces blood-brain barrier (BBB) function and neurological recovery after stroke.
(A) Image showing the leakage of Evans blue in Cdh5CreERT2 injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus at RP34D after middle cerebral artery occlusion (MCAO) and quantitative analysis of the leakage of Evans blue (n = 5). (B) Immunofluorescence staining of NeuN expression in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after MCAO at RP34D and quantitative analysis of the number in the unit area (n = 3, 20 slices/mouse). (C) Graph showing rotarod test in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after MCAO (n = 8–10). (D) Graph showing beam walking test in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRedWPREs virus after MCAO (n = 8–10). (E) Graph showing corner test in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after MCAO (n = 8–10). (F) Graph showing adhesive movement test in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after MCAO (n = 8–10). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A-F).
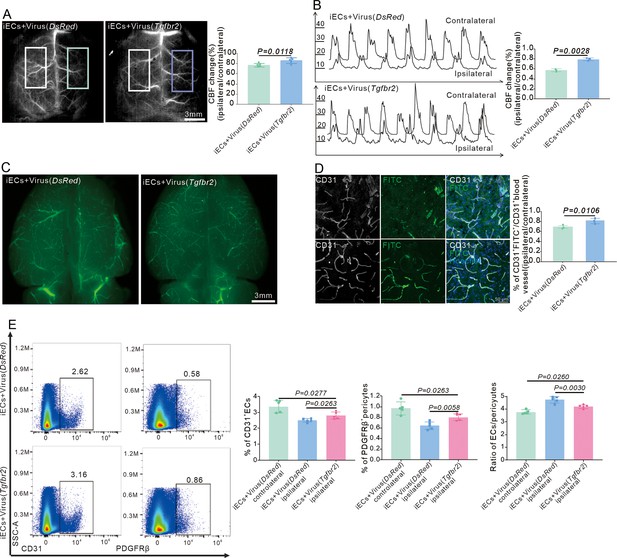
Endothelial cell (EC)-specific overexpression of the Tgfbr2 gene reinforces cerebral blood flow (CBF) and vessel length.
(A) Laser speckle image showing the change of CBF in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after middle cerebral artery occlusion (MCAO) at RP34D and quantitative analysis of the change of CBF (n = 5). (B) Graph showing the change of CBF using Doppler test in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after MCAO at RP34D and quantitative analysis of the change percentage (n = 5). (C) Image showing FITC-Dextran in gelatin from Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after MCAO at RP34D and quantitative analysis of the percentage (n = 3). (D) Immunofluorescence staining of CD31 expression in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after MCAO at RP34D and quantitative analysis of the percentage, FITC-Dextran in gelatin injection by cardiac perfusion (n = 3, 20 slices/mouse). (E) Flow cytometry analysis of the proportion of PDGFR+ cells and CD31+ cells in Cdh5CreERT2 mice injected with AAV2/9-BI30-EF1α-DIO-Tgfbr2-3XFLAG-P2A-DsRed-WPREs virus after MCAO at RP34D and quantitative analysis of the percentage (n = 5). Data are presented as mean ± SEM, unpaired two-tailed Student’s t-test (A-B and D-E).
Tables
| Markers | ECs | Fibroblast-like cell (RP8D) | Pericyte (RP34D) |
|---|---|---|---|
| CD31 | ✓ | xx | xx |
| GLUT1 | ✓ | xx | xx |
| ERG | ✓ | xx | xx |
| VE-Cadherin | ✓ | xx | xx |
| PDGFRa | xx | ✓ | xx |
| PDGFR beta | xx | ✓ | sqrt() |
| Vimentin | xx | ✓ | xx |
| CD13 | xx | xx | ✓ |
| NG2 | xx | xx | sqrt() |