Viral-mediated Pou5f1 (Oct4) overexpression and inhibition of Notch signaling synergistically induce neurogenic competence in mammalian Müller glia
Figures
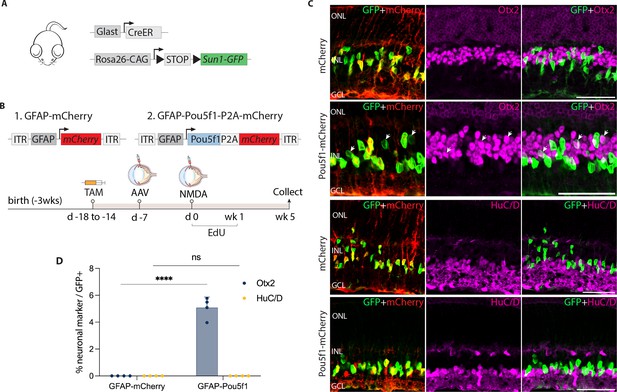
AAV-mediated overexpression of Pou5f1 induces neurogenesis in control GlastCreER;Rosa26LSL-Sun1-GFP Müller glia following N-methyl-D-aspartate (NMDA)-induced excitotoxicity.
(A) Schematic of the transgenic construct used to specifically label Müller glia with Sun1-GFP expression. (B) Schematic of the GFAP AAV constructs and experimental workflow. (C) Representative images of retinas immunolabeled for GFP, mCherry, Otx2, and HuC/D. White arrowheads indicate GFP-positive Müller glia-derived neurons expressing neuronal markers Otx2 or HuC/D. (D) Quantification of mean percentage ± SD of GFP-positive Müller glia-derived neurons expressing either Otx2 or HuC/D. Significance was determined via two-way ANOVA with Tukey’s multiple comparison test: ****p < 0.0001. Each data point was calculated from an individual retina. TAM, tamoxifen; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 μm.
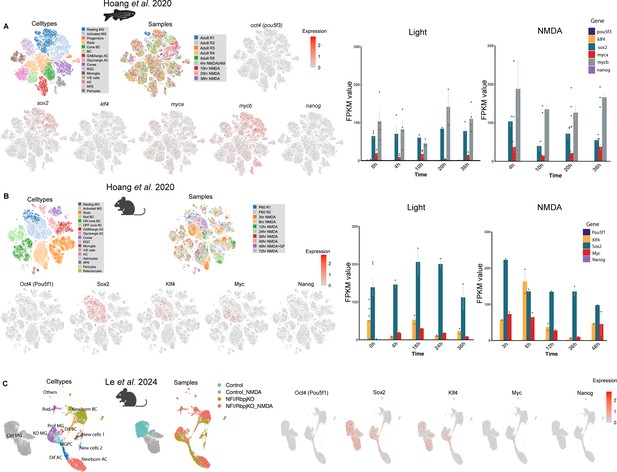
Oct4/pou5f3 mRNA is not detectably expressed in Müller glia after injury.
Expression of pluripotency factors in resting or injured retinas by single-cell RNA-sequencing (scRNA-seq) and bulk RNA data in (A) zebrafish and (B) mouse after retinal injury from Hoang et al., 2020 and in (C) Nfia/b/x;Rbpj-deficient, Sun1-GFP-positive Müller glia from Le et al., 2024.
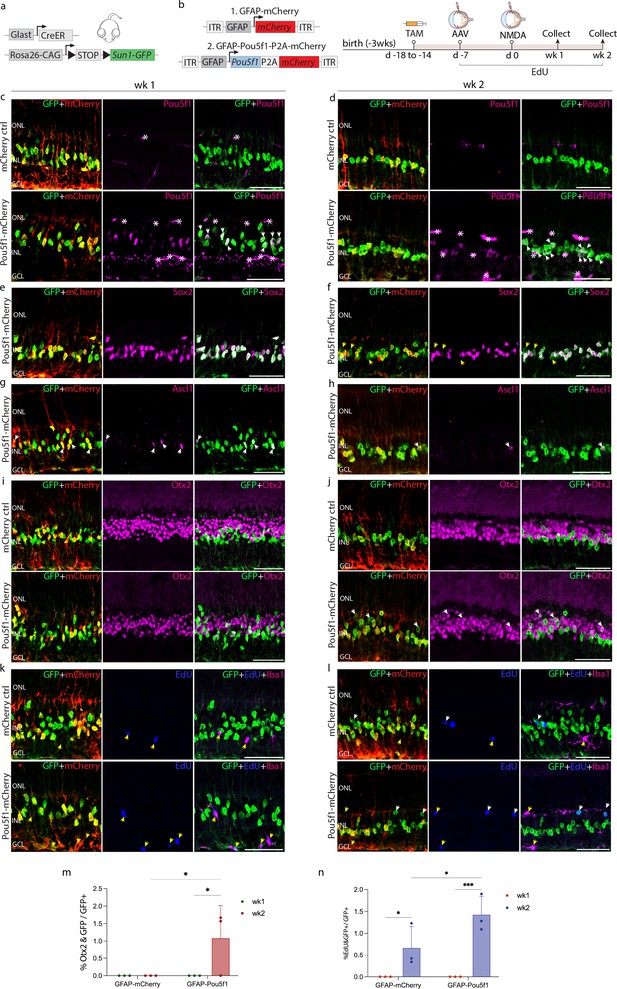
Immunohistochemical analysis of glial markers, AAV reporter expression, and other molecular markers at 1 and 2 weeks following N-methyl-D-aspartate (NMDA) in control GlastCreER;Rosa26LSL-Sun1-GFP Müller glia.
(a) Schematic of the transgenic construct used to specifically label Müller glia with Sun1-GFP expression. (b) Schematic of the GFAP AAV constructs and experimental workflow. Representative images of retinas immunolabeled for GFP, mCherry and (c, d) Pou5f1, (e, f) Sox2, (g, h) Ascl1, (i, j) Otx2, and (k, l) EdU/Iba1. White arrowheads indicate GFP-positive cells expressing selected markers. Yellow arrowheads indicate EdU-positive cells that do not co-label with Sox2, indicating cells that have undergone Müller glia to bipolar cell conversion, and EdU/Iba1+ cells that do not co-label with GFP+ cells. Asterisks (*) indicate mouse-on-mouse vascular staining. Quantification of mean percentage ± SD of GFP-positive Müller glia co-labeled with (m) Otx2 and (n) EdU. Significance was determined via two-way ANOVA with Tukey’s multiple comparison test: ***p < 0.001, *p < 0.05. Each data point was calculated from an individual retina. TAM, tamoxifen; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 μm.
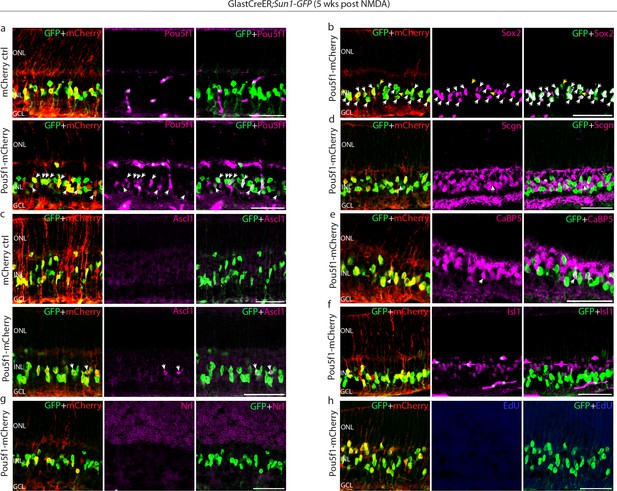
Immunohistochemical analysis of glial markers, AAV reporter expression, and other molecular markers at 5 weeks following N-methyl-D-aspartate (NMDA) in control Müller glia.
Representative images of retinas from control GlastCreER;Rosa26LSL-Sun1-GFP mice immunolabeled for GFP, mCherry, and (a) Pou5f1, (b) Sox2, (c) Ascl1, (d) Scgn, (e) Cabp5, (f) Isl1, (g) Nrl, and (h) EdU. White arrowheads indicate GFP-positive cells co-labeled with the relevant marker. Yellow arrowheads indicate GFP-positive Müller glia that did not co-labeled with Müller glia markers. Asterisks (*) indicate mouse-on-mouse vascular staining. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 μm.
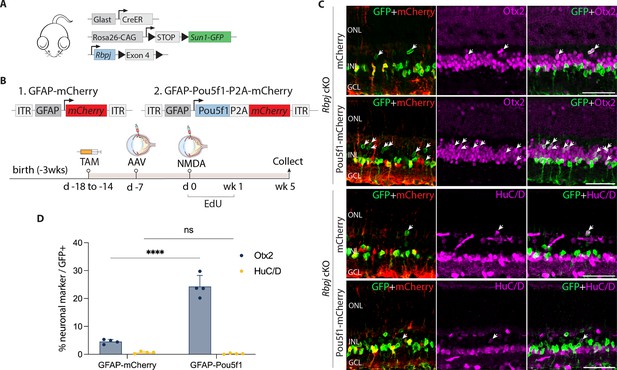
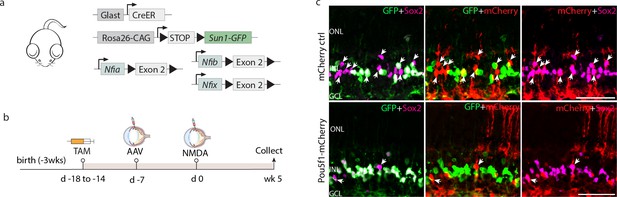
Pou5f1 overexpression enhances neurogenesis in Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia.
(A) Schematic of the transgenic constructs used to induce loss of function of Rbpj specifically in Müller glia. (B) Schematic of the GFAP AAV constructs and experimental workflow. (C) Representative images of retinas immunolabeled for GFP, mCherry, Otx2, and HuC/D. White arrowheads indicate GFP-positive Müller glia-derived neurons expressing neuronal markers Otx2 or HuC/D. (D) Quantification of mean percentage ± SD of GFP-positive Müller glia-derived neurons expressing either Otx2 or HuC/D. Significance was determined via two-way ANOVA with Tukey’s multiple comparison test: ****p < 0.0001. TAM, tamoxifen; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 μm.
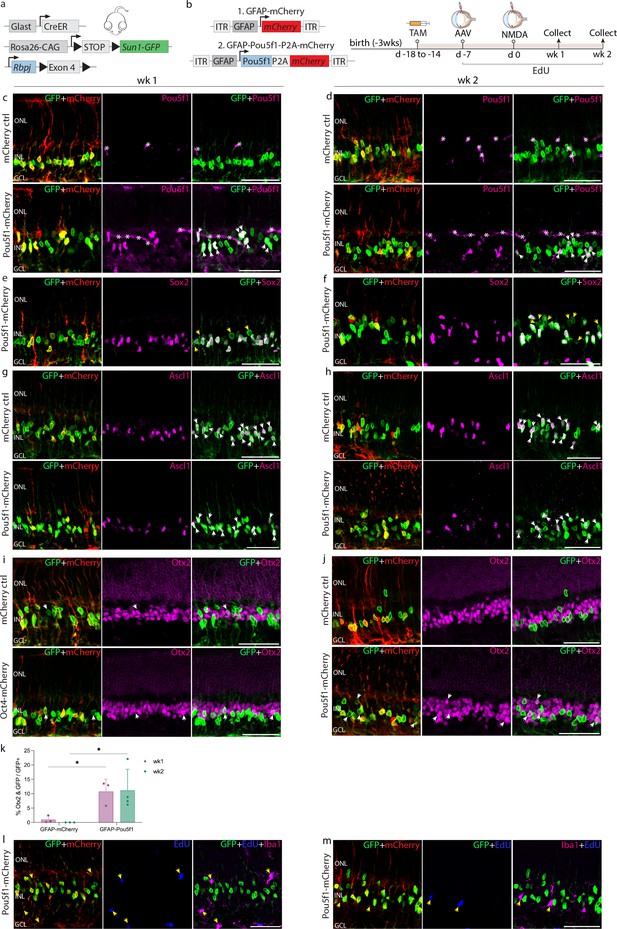
Immunohistochemical analysis of glial markers, AAV reporter expression, and other molecular markers at 1 and 2 weeks following N-methyl-D-aspartate (NMDA) in Rbpj-deficient Müller glia.
(a) Schematic of the transgenic constructs (GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP) used to induce loss of function of Rbpj specifically in Müller glia. (b) Schematic of the GFAP AAV constructs and experimental workflow. Representative images of retinas immunolabeled for GFP, mCherry and (c, d) Pou5f1, (e, f) Sox2, (g, h) Ascl1, (i, j) Otx2 and (l, m) EdU/Iba1. White arrowheads indicate GFP-positive cells expressing selected markers. Yellow arrowheads indicate GFP-positive cells that do not co-label with Sox2, and EdU/Iba1+ cells that do not express GFP. Asterisks (*) indicate mouse-on-mouse vascular staining. (k) Quantification of mean percentage ± SD of GFP-positive Müller glia-derived neurons expressing Otx2. Significance was determined via two-way ANOVA with Tukey’s multiple comparison test: *p < 0.05. Each data point was calculated from an individual retina. TAM, tamoxifen; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 μm.
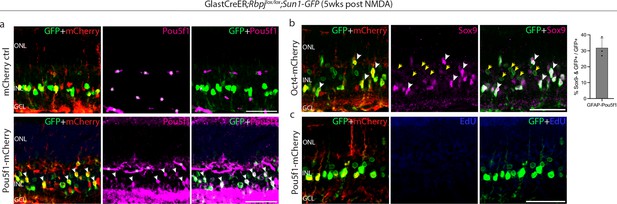
Immunohistochemical analysis of glial markers, AAV reporter expression, and other molecular markers at 5 weeks following N-methyl-D-aspartate (NMDA) in Rbpj-deficient Müller glia.
Representative images of retina from GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP mice immunolabeled for GFP, mCherry, and (a) Pou5f1, (b) Sox9, and (c) EdU. White arrowheads indicate GFP-positive cells co-labeled with the relevant marker. Yellow arrowheads indicate GFP-positive Müller glia that did not co-labeled with Müller glia markers. Asterisks (*) indicate mouse-on-mouse vascular staining. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 μm. (b) Quantification of mean percentage ± SD of GFP-positive Müller glia without Sox9 expression.
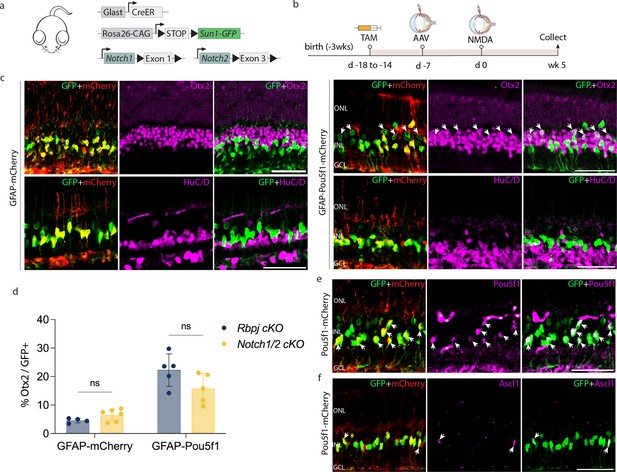
AAV-mediated overexpression of Pou5f1 induces neurogenesis in injured Notch1/2-deficient Müller glia.
(a) Schematic of the transgenic mice (GlastCreER;Notch1lox/lox;Notch2lox/lox;Rosa26LSL-Sun1-GFP) used to induce deletion of Notch1/2 specifically in Müller glia. (b) Schematic of the experimental workflow. Representative images of retinas immunolabeled for GFP, mCherry, and (c) Otx2, HuC/D, (e) Pou5f1, and (f) Ascl1. White arrowheads indicate GFP-positive Müller glia expressing Otx2, Pou5f1, or Ascl1. Asterisks (*) indicate mouse-on-mouse vascular staining. (d) Quantification of mean percentage ± SD of GFP-positive Müller glia-derived neurons expressing either Otx2 or HuC/D. Significance was determined via two-way ANOVA with Tukey’s multiple comparison test: ns=not significant. TAM, tamoxifen; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 μm.
Pou5f1 overexpression does not enhance neurogenesis in Nfia/b/x-deficient Müller glia because the GFAP minipromoter is not active in these cells.
(a) Schematic of the transgenic mice (GlastCreER;Nfialox/lox;Nfiablox/lox;Nfialox/lox;Rosa26LSL-Sun1-GFP) used to induce deletion of Nfia/b/x specifically in Müller glia. (b) Schematic of the experimental workflow. (c) Representative images of retinas immunolabeled for GFP, mCherry, and Sox2. White arrowheads indicate mCherry expression in Sox2+ Müller glia that are not Sun1-GFP+. Quantification of mean percentage ± SD of GFP-positive Müller glia-derived neurons expressing either Otx2 or HuC/D. TAM, tamoxifen; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 μm.
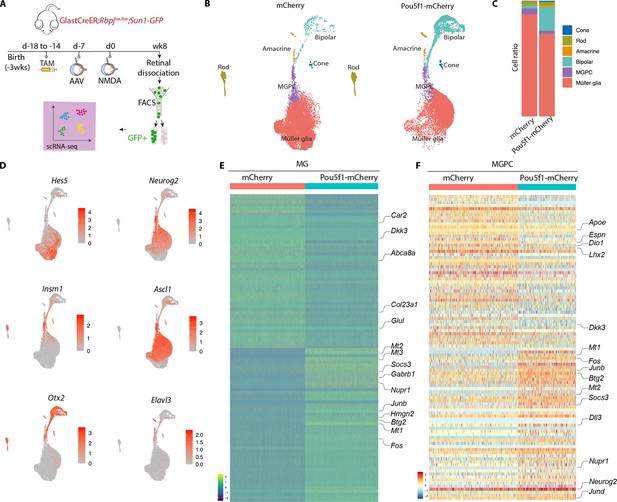
Single-cell RNA-sequencing (scRNA-seq) analysis of Müller glia and Müller glia-derived neurons from Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia following Pou5f1 overexpression.
(A) Schematic of the scRNA-seq experimental pipeline. (B) UMAP plot showing the clustering of GFP-positive cells from Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia retinas infected with GFAP-mCherry and GFAP-Pou5f1-mCherry AAV constructs. (C) Stacked bar plots showing the proportion of cells in each cluster across two sample groups. (D) Feature plots highlighting the cluster of Müller glia (Hes5), neurogenic Müller glia-derived progenitor cells (MGPCs) (Neurog2, Insm1, Ascl1), bipolar cells (Otx2), and amacrine cells (Elavl3). (E) Heatmap showing the expression of top differentially expressed genes (DEGs) from the Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia cell cluster from retinas infected with GFAP-mCherry and GFAP-Pou5f1-mCherry AAV constructs. (F) Heatmap showing the expression of top DEGs for MGPC cell cluster from retinas infected with GFAP-mCherry and GFAP-Pou5f1-mCherry AAV constructs.
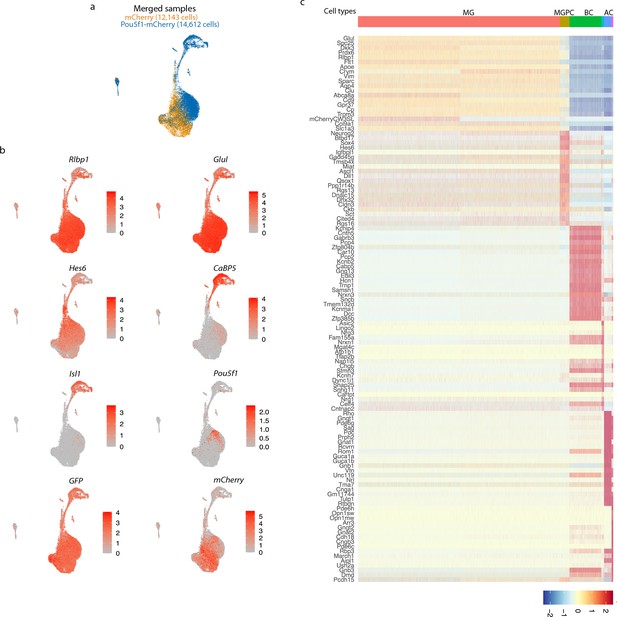
Single-cell RNA-sequencing (scRNA-seq) analysis of Müller glia-derived neurons from Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia following Pou5f1 overexpression.
(a) UMAP plot showing GFP-positive cells from retinas infected with GFAP-mCherry and GFAP-Pou5f1-mCherry AAVs. (b) Expression of glial and neuronal markers. (c) Top differentially expressed genes (DEGs) of different cell types generated by Pou5f1 overexpression in Rbp-deficient Müller glia.
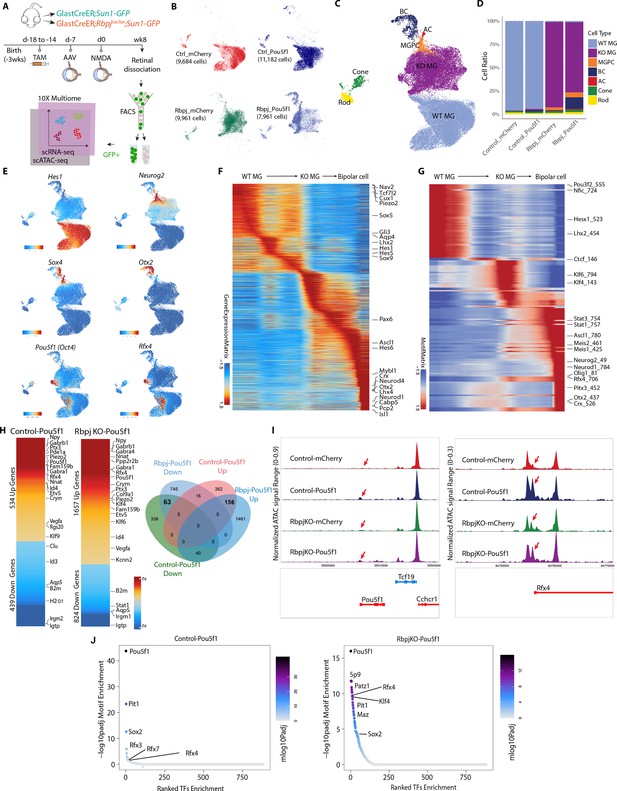
Integrated snRNA/scATAC-seq analysis of control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia following Pou5f1 overexpression.
(A) Schematic of the multiomic scRNA/ATAC-seq experimental pipeline. (B) UMAP plot of multiomic datasets showing the clustering of GFP+ cells from control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia from GFAP-mCherry and GFAP-Pou5f1-mCherry AAV infected retinas. (C) UMAP plot showing the identity of cell clusters determined by marker gene expression. (D) Stacked bar plots represent the proportion of cells in each cluster across different sample groups. (E) Feature plots highlighting the cluster of Müller glia (Hes1), neurogenic Müller glia-derived progenitor cells (MGPCs) (Neurog2, Sox4), bipolar cells (Otx2), Pou5f1- and Rfx4-expressing cells. (F) Heatmap showing expression of differentially expressed genes (DEGs) along the neurogenesis trajectory. (G) Heatmap showing differential motif activity along the neurogenesis trajectory. (H) Heatmaps of DEGs differentially expressed between Pou5f1 and mCherry control samples in the control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia clusters. Venn diagram showing unique and common DEGs between Pou5f1 and mCherry control in the control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia. (I) Increased chromatin accessibility regions associated with the Pou5f1 and Rfx4 loci observed following Pou5f1 overexpression in both control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia. (J) Top-ranked enriched motifs in chromatin regions showing increased accessibility following Pou5f1 overexpression in both control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia cell clusters.
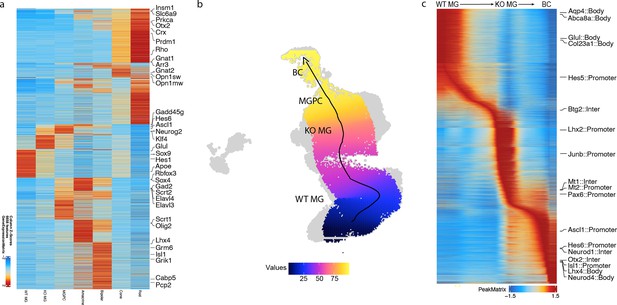
Pseudotime analysis showing the transition from resting to neurogenic Müller glia-derived progenitor cells (MGPCs) to bipolar cells.
(a) Top differentially expressed genes (DEGs) across different retinal cell types. (b) Pseudotime trajectory of neurogenesis from Müller glia to bipolar cells. (c) Heatmap showing differential chromatin accessible regions at different pseudotime stages along the neurogenesis trajectory.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Chicken anti-GFP (chicken polyclonal) | ThermoFisher | A10262, RRID:AB_2534023 | IF (1:400) |
| Antibody | Rabbit anti-RFP (rabbit polyclonal) | Abcam | ab124754, RRID:AB_10971665 | IF (1:400) |
| Antibody | Goat anti-RFP (goat polyclonal) | Rockland | 200-101-379, RRID:AB_2744552 | IF (1:400) |
| Antibody | Goat anti-Otx2 (goat polyclonal) | R&D Systems | AF1979, RRID:AB_2157172 | IF (1:200) |
| Antibody | Mouse anti-HuC/D (mouse monoclonal) | ThermoFisher | A-21271, RRID:AB_221448 | IF (1:200) |
| Antibody | Mouse anti-NeuN (mouse monoclonal) | Sigma-Aldrich | MAB377, RRID:AB_2298772 | IF (1:200) |
| Antibody | Mouse anti-Oct3/4 (mouse monoclonal) | Santa Cruz | sc-5279, RRID:AB_628051 | IF (1:200) |
| Antibody | Rabbit anti-Sox9 (mouse monoclonal) | Sigma-Aldrich | AB5535, RRID:AB_2239761 | IF (1:400) |
| Antibody | Goat anti-Nrl (goat polyclonal) | R&D Systems | AF2945, RRID:AB_2155098 | IF (1:400) |
| Antibody | Rabbit anti-Scgn (rabbit polyclonal) | Biovendor Laboratory Medicine | RD181120100, RRID:AB_2034060 | IF (1:400) |
| Antibody | Rabbit anti-Ascl1 (rabbit monoclonal) | Abcam | ab211327, RRID:AB_2924270 | IF (1:400) |
| Antibody | Rabbit anti-CaBP5 (rabbit monoclonal) | SynapticSystems | 475 002, RRID:AB_2924962 | IF (1:400) |
| Antibody | Goat anti-Sox2 (goat polyclonal) | R&D Systems | AF2018, RRID:AB_355110 | IF (1:400) |
| Antibody | Mouse anti-Isl1 (mouse monoclonal) | DSHB | 40.2D6, RRID:AB_528315 | IF (1:400) |
| Antibody | Donkey anti-Chicken 488 (donkey polyclonal) | Sigma-Aldrich | SAB4600031, RRID:AB_2721061 | IF (1:400) |
| Antibody | Donkey anti-Rabbit 568 (donkey polyclonal) | ThermoFisher | A-10042, RRID:AB_2757564 | IF (1:400) |
| Antibody | Donkey anti-Goat 568 (donkey polyclonal) | ThermoFisher | A11057, RRID:AB_2534104 | IF (1:400) |
| Antibody | Donkey anti-Mouse 568 (donkey polyclonal) | ThermoFisher | A10037, RRID:AB_2534013 | IF (1:400) |
| Antibody | Donkey anti-Rat 568 (donkey polyclonal) | ThermoFisher | A78946, RRID:AB_2910653 | IF (1:400) |
| Antibody | Donkey anti-Goat 633 (donkey polyclonal) | ThermoFisher | A-21082, RRID:AB_2535739 | IF (1:400) |
| Antibody | Donkey anti-Rabbit 647 (donkey polyclonal) | ThermoFisher | A-31573, RRID:AB_2536183 | IF (1:400) |
| Antibody | Donkey anti-Mouse 647 (donkey polyclonal) | ThermoFisher | A-31571, RRID:AB_162542 | IF (1:400) |
| Commercial assay or kit | 10 x scRNaseq 3′ v3.1 | 10 X Genomics | 1000268 | |
| Commercial assay or kit | 10 x Multiome ATAC +GEX | 10 X Genomics | 1000283 | |
| Commercial assay or kit | Click-iT EdU Alexa Fluor 647 | ThermoFisher | C10340 | |
| Commercial assay or kit | Click-iT EdU Pacific Blue | ThermoFisher | C10418 | |
| Software, algorithm | ImageJ/Fiji | https://imagej.net/software/fiji/ | RRID:SCR_002285 | |
| Software, algorithm | Adobe Illustrator | http://www.adobe.com | RRID:SCR_010279 | v.26.5 |
| Software, algorithm | GraphPad Prism | https://www.graphpad.com/ | RRID:SCR_002798 | v.10 |
| Software, algorithm | Cell Ranger | 10 X Genomics | RRID:SCR_017344 | v.2.0.2 |
| Software, algorithm | ArchR | https://github.com/GreenleafLab/ArchR | RRID:SCR_020982 | v.1.0.2 |
| Software, algorithm | Seurat | https://github.com/satijalab/seurat | RRID:SCR_007322 | v.5.1.0 |
Additional files
-
Supplementary file 1
ScRNA-seq analysis of Sun1-GFP Müller glia-derived cells identifies genes differentially regulated by Pou5f1 overexpression.
Raw + adjusted p-values, relative fold-change, and the fraction of all profiled cells expressing the gene in question is shown for both Muller glia and Muller glia-derived progenitor cells transfected with AAV-GFAP-Pou5f1-mCherry vs. AAV-GFAP-mCherry.
- https://cdn.elifesciences.org/articles/106450/elife-106450-supp1-v1.xls
-
Supplementary file 2
Multiomic analysis identifies differentially expressed genes (DEGs), differential chromatin accessible regions, and differentially accessible transcription factor target motifs are shown at discrete pseudotime intervals along the Müller glia-bipolar differentiation trajectory.
- https://cdn.elifesciences.org/articles/106450/elife-106450-supp2-v1.xls
-
Supplementary file 3
SnRNA-seq analysis identifies differentially expressed genes (DEGs) induced by Pou5f1 overexpression from control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia cell clusters.
- https://cdn.elifesciences.org/articles/106450/elife-106450-supp3-v1.xlsx
-
Supplementary file 4
SnATAC-seq analysis identifies differentially accessible chromatin regions regulated by Pou5f1 overexpression in control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia cell clusters.
- https://cdn.elifesciences.org/articles/106450/elife-106450-supp4-v1.xls
-
Supplementary file 5
SnATAC-seq analysis identifies differential motif activity in increased accessible chromatin regions by Pou5f1 overexpression in control GlastCreER;Rosa26LSL-Sun1-GFP and Rbpj-deficient GlastCreER;Rbpjlox/lox;Rosa26LSL-Sun1-GFP Müller glia cell clusters.
- https://cdn.elifesciences.org/articles/106450/elife-106450-supp5-v1.xls
-
Supplementary file 6
List of AAV constructs used in the study.
Columns identify the AAV construct, titer, and injection volume.
- https://cdn.elifesciences.org/articles/106450/elife-106450-supp6-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/106450/elife-106450-mdarchecklist1-v1.pdf





