NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation
Figures
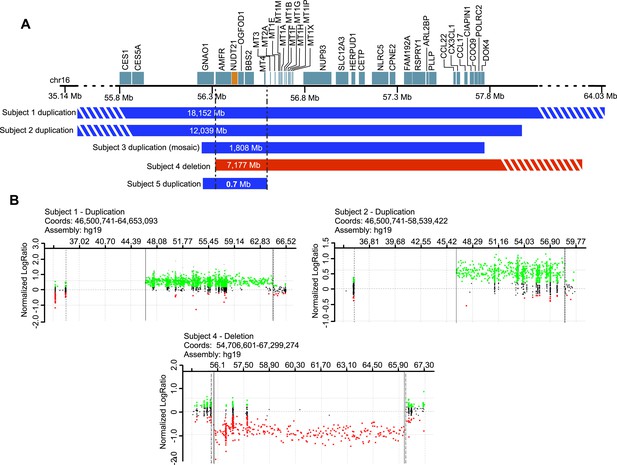
Subjects with NUDT21-spanning copy-number variations (CNVs).
(A) Five intrachromosomal rearrangements of chromosome 16q including the NUDT21 gene, identified by clinical array comparative genomic hybridization. Duplications shown in blue, deletion in red. Del, deletion; Dup, duplication; Mb, megabases. The striped bars indicate that the copy variant is not drawn to scale. (B) Array plots of oligonucleotide arrays on subjects 1, 2, and 4. The array plots of subject 3 and 5 were unavailable due to the closure of the Signature Genomics microarray laboratory in June 2014. Black dots indicate probes with normal copy-number, green dots indicate copy-number gain, and red dots indicate copy-number loss. Solid and dotted lines respectively define the minimum and maximum expected boundaries of the CNVs.
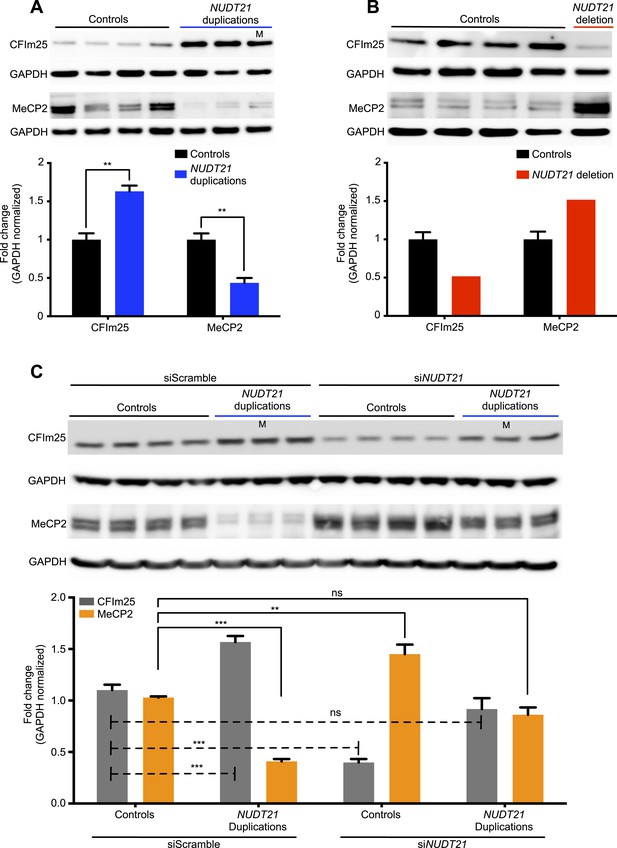
CFIm25 regulates MeCP2 protein levels in patient-derived lymphoblastoid cells with NUDT21 CNVs.
(A) Representative western blot picture for three duplication patients compared to four age-matched controls showing the increase of CFIm25 and decrease of MeCP2 protein levels. (B) Representative western blot picture for one deletion patient compared to four age-matched controls showing the decrease of CFIm25 and increase of MeCP2 protein levels. Quantification of protein levels for both CFIm25 and MeCP2 from three duplication patients and one deletion patient compared to a total of 13 age-matched controls are shown below the corresponding western blot. Data represent mean ± SEM from a total of six technical replicates. Data were normalized to GAPDH protein levels. (C) Western blot and its relative quantification showing that knockdown of NUDT21 by siRNA-NUDT21 nucleofection increases MeCP2 in control and duplication subjects, and normalizing CFIm25 in duplication patients rescue MeCP2 to control levels. Data represent mean ± SEM from four age-matched control and three duplication cases. Data were normalized to GAPDH protein levels. **p < 0.01, ***p < 0.001. M, mosaic patient.
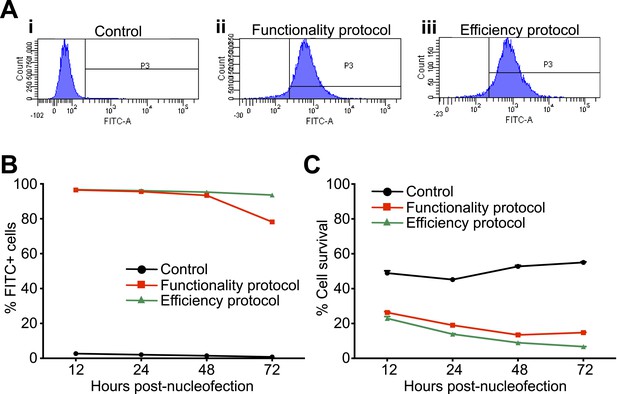
siGLO nucleofection showing patient-derived lymphoblastoid cells can be transfected with small RNA.
(A) Representative histograms of control (i) and siGLO-nucleofected lymphoblastoid cells showing nearly 100% efficiency using both the functionality (ii) and efficiency (iii) protocols. (B) Time series showing the presence of small RNA in nearly 100% of nucleofected lymphoblastoid cells up to 48 hr after nucleofection. (C) Time series showing cell survival following nucleofection using either the functionality or efficiency nucleofection protocol.
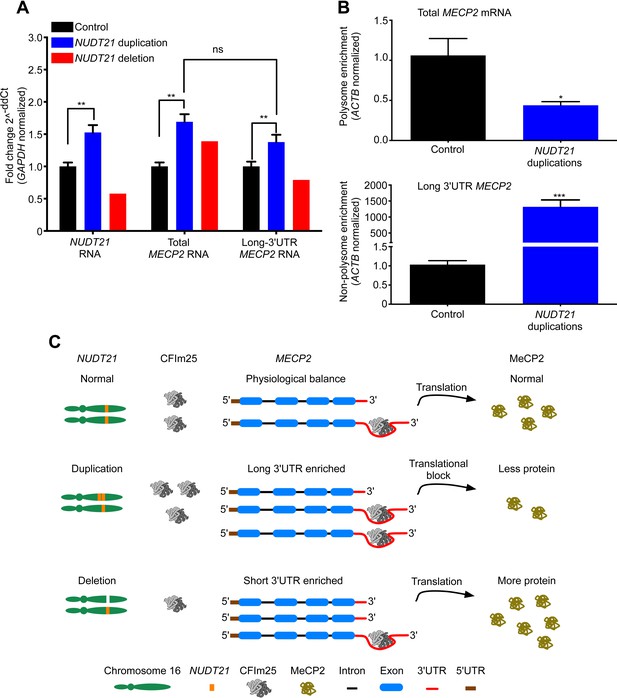
NUDT21 mRNA levels correlate with inefficiently translated long MECP2 mRNA.
(A) RNA quantification by quantitative RT-polymerase chain reaction (qRT-PCR) from lymphoblastoid cells of NUDT21 duplication and deletion patients. The bar graph shows the total mRNA fold change of NUDT21, total MECP2, and long MECP2 for the three duplication patients and one deletion patient compared to 13 age-matched controls. Data represent mean ± SEM from five independent experiments. Data were normalized to GAPDH mRNA levels. (B) Relative polyribosomal and non-poyribosomal enrichment of total and long MECP2 mRNA isoforms of NUDT21 duplication patients compared to age-matched controls. Data represent mean ± SEM from a total of three control and duplication cases. Data were normalized to ACTB mRNA levels. (C) Proposed model showing that duplication and deletion patients have more or less CFIm25, respectively leading to a relative increase in long and short MECP2 3′ UTR isoforms. In both cases, there is an accumulation of mRNA: in the deletion patient, this leads to more MeCP2 protein, but in the duplication patients, it results in less MeCP2 protein due to a translational block from the CFIm25-mediated increase in long MECP2 isoforms and putative binding of miRNAs or RNA-binding proteins to the 3′ UTR. *p < 0.05, **p < 0.01, ***p < 0.001.
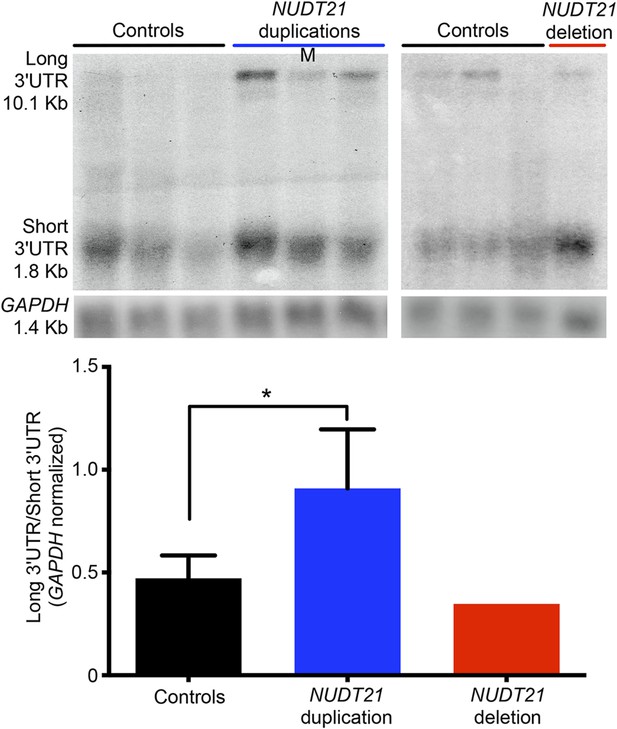
Northern blot assay from patient-derived lymphoblastoid cells.
Northern blot assay of lymphoblastoid cells showing that duplication (left panel) and deletion (right panel) patients respectively have more long or short MECP2 3′ UTR isoforms. Relative northern blot quantification of three duplication patients and one deletion patient compared to 13 age-matched controls (bottom panel). Data represent mean ± SEM from four independent experiments. Data were normalized to GAPDH mRNA levels. For all the experiments, p values were calculated by Student's t-test comparing controls with duplication patients. *p < 0.05, **p < 0.01. M, mosaic patient.
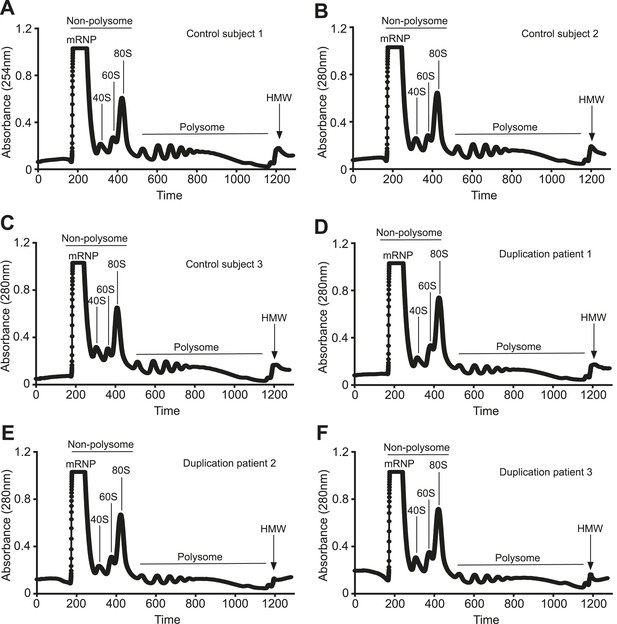
Polyribosome fractionation traces of control and duplication subjects.
Representative polyribosome traces from control (A-C) and duplication (D-F) subjects. UV absorption at 254 nm was plotted vs time depicting the successful resolution of the different ribosomal fractions.
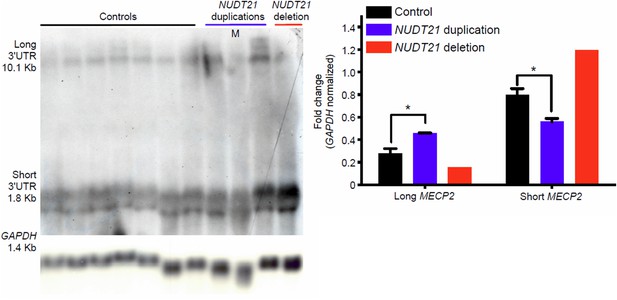
Northern blot assay from patient-derived lymphoblast cells.
Northern blot assay showing that duplication and deletion patients respectively have more long or short MECP2 isoforms (left panel). Quantification of three duplication patients and one deletion compared to 7 age-matched controls (right panel). Data represent mean ± SEM. The p values were calculated by Student's t-test comparing controls with duplication patients. *p<0.05. M=mosaic patient.
Tables
Molecular and clinical characteristics of four individuals with NUDT21 duplication and one individual with NUDT21 deletion
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | Subject 5 | |
|---|---|---|---|---|---|
| Sex | F | M | M | F | F |
| Age | 10 years | 5 years | 15 years | 13 years | 8 years |
| Deletion/duplication | Duplication | Duplication | Duplication | Deletion | Duplication |
| Coordinates | chr16: 46,500,741-64,653,093 | chr16: 46,500,741-58,539,422 | chr16: 55,725,264-57,533,101 | chr16: 56,344,856-63,521,523 | chr16: 55,919,145-56,619,283 |
| Size | 18.15 Mb | 12.04 Mb | 1.81 Mb | 7.18 Mb | 0.70 Mb |
| Zygosity | Heterozygous | Heterozygous | Mosaic marker chromosome (present in 45% of cells) | Heterozygous | Heterozygous |
| Inheritance | De novo | De novo | De novo | Unknown | Not maternal* |
| Additional CNVs | 52 kb duplication of chr6: 152, 716, 341-152, 768, 421 (maternal) | None | None | 144 kb duplication of chr17: 427, 284-571, 275 (inheritance unknown) | None |
| Developmental delay/intellectual disability | Intellectual disability (clinical impression: moderate) | Significant developmental delay | Intellectual disability | Intellectual disability (full scale IQ 53) | Intellectual disability |
| Developmental regression | Yes, at 2 years of age | No | Yes | No | No |
| Autism spectrum disorder | Yes | No | Yes | No | Yes |
| Epilepsy | Single, isolated seizure at 5 years of age | No | No | Intractable, symptomatic partial onset seizures | No |
| ADHD | Yes | Yes | Unknown | Yes | No |
-
F, female; M, male; chr, chromosome; MB, megabases; CNVs, copy number variants; kb, kilobases; ADHD, attention deficit hyperactivity disorder.
-
*
Father not available.





