Non-canonical antagonism of PI3K by the kinase Itpkb delays thymocyte β-selection and renders it Notch-dependent
Figures
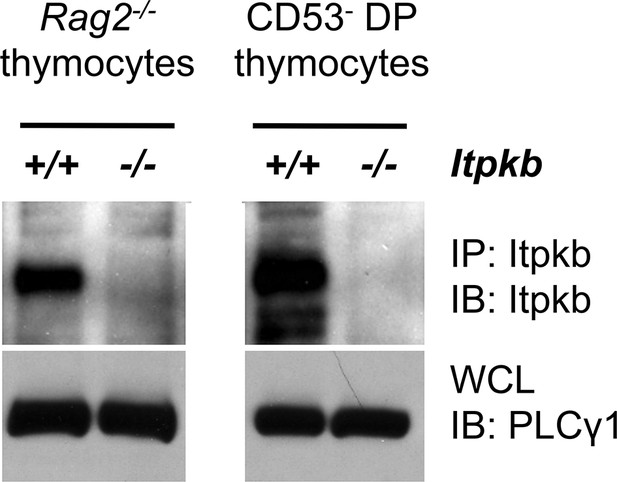
Itpkb protein is expressed in DN3 cells from Itpkb+/+ but not Itpkb-/- mice.
Shown are immunoblots (IB) of Itpkb immunoprecipitates (IP, top) or whole cell lysates (WCL, bottom) from Itpkb+/+ or Itpkb-/- DN3 cell-enriched Rag2-/- thymocytes (left) or sorted CD53- DP thymocytes (right), resolved via SDS-PAGE and probed with antibodies against Itpkb (top) or PLCγ1 (bottom, loading control) as in (Miller et al., 2007; Huang et al., 2007).
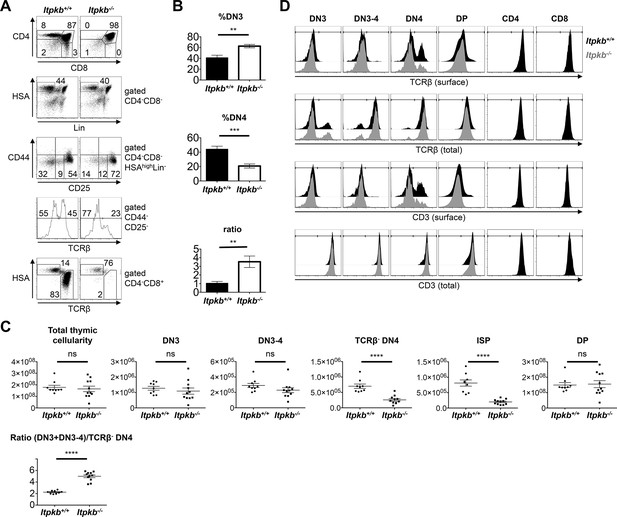
Altered β-selection in Itpkb-/- mice.
(A) Flow cytometric profiles of thymocytes from Itpkb+/+ and Itpkb-/- littermate mice. Top, CD4 and CD8 expression. Upper center, HSA and mature lineage marker (Lin = CD11b, CD11c, CD19, B220, CD49b, Gr-1, Ter119, TCRγ) expression on CD4-CD8- (DN) cells. Lower center, CD44 and CD25 expression on HSAhighLin- DN cells. The bottom gates denote DN3, transitional DN3-4 and DN4 cells from right to left. Bottom two panels, TCRβ expression on DN4 cells, and HSA and TCRβ expression on CD4-CD8+ cells with ISP (HSAhighTCRβlow) and mature CD8 T cell (HSAlowTCRβhigh) gates. Numbers denote % cells per gate. Representative of at least 7 independent experiments. (B) Mean ± SEM %DN3 cells, %DN4 cells or %DN3:%DN4 cell ratio in Itpkb+/+ or Itpkb-/- 5–7 week old littermates. Statistical significance of genotype differences was analyzed by unpaired two-tailed Student's t-tests (n = 7). (C) Total numbers of thymocytes, Lin-HSAhigh CD44-CD25+ DN3, CD44-CD25int DN3-4, TCRβ-CD44-CD25- DN4, CD8α+HSAhighTCRβlow ISP or CD4+CD8+ DP cells in individual Itpkb+/+ or Itpkb-/- mice. Horizontal lines denote means ± SEM. Significance of genotype differences was analyzed as in (B). nWT = 9, nItpkb-/- = 11. ns, no significant difference. (D) Histograms of surface or total cellular TCRβ or CD3 levels on the indicated thymocyte populations from Itpkb+/+ (black) or Itpkb-/- (gray) mice. Representative of ≥3 independent experiments.
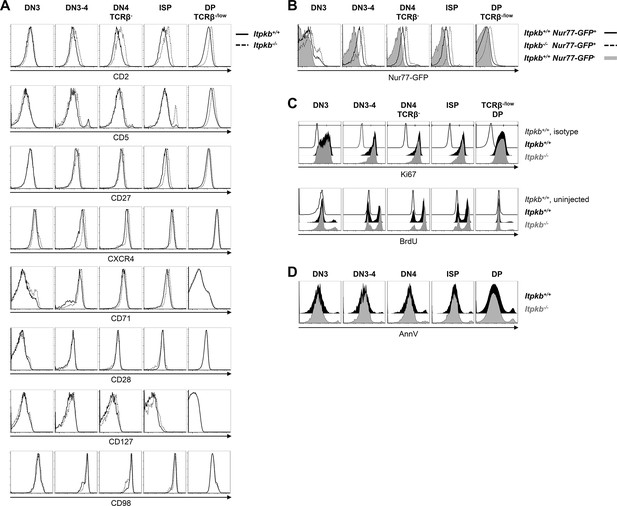
Surface marker expression, steady-state proliferation and viability of Itpkb+/+ and Itpkb-/- thymocytes.
(A) Surface levels of the indicated markers for activation or β-selection on thymocyte subpopulations from Itpkb+/+ (solid) or Itpkb-/- (hatched) mice. (B) Nr4a1/Nur77-GFP expression in Itpkb+/+ or Itpkb-/- Nr4a1/Nur77-GFP transgenic (solid or hatched black, respectively) or non-transgenic (gray) mice. Representative of ≥3 independent experiments. (C) Steady-state proliferative status of the indicated thymocyte subpopulations in Itpkb+/+ (black) or Itpkb-/- (gray) mice was analyzed by Ki67 stain (top, representative of 3 independent experiments) or BrdU incorporation assay (bottom, representative of 2 independent experiments). Thin open histograms, Itpkb+/+ isotype or BrdU-uninjected, respectively, negative control. TCRβlow DP cells were analyzed as they represent the majority of DP cells and Itpkb-/- mice lack TCRβhigh DP cells (Wen et al., 2004). (D) Steady-state viability of the indicated thymocyte subpopulations in Itpkb+/+ (black) or Itpkb-/- (gray) mice was analyzed by AnnexinV (AnnV) stain. Representative of 4 independent experiments.
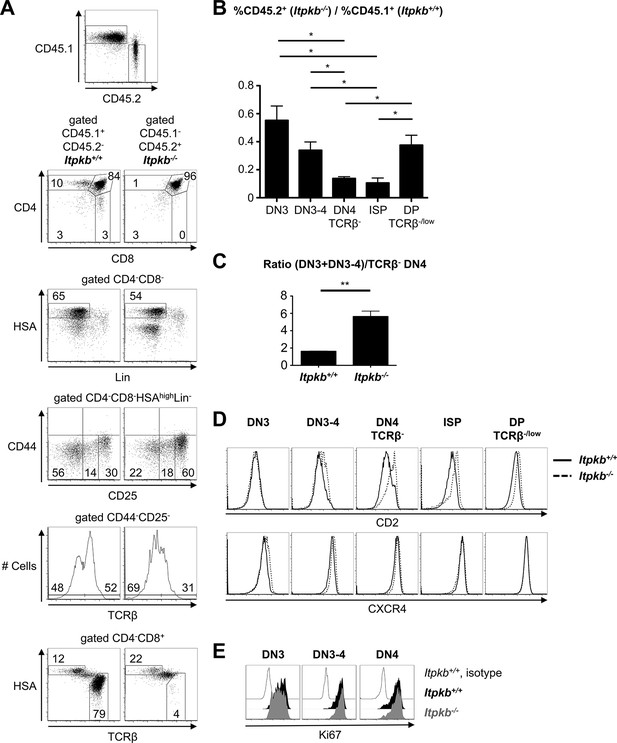
Itpkb controls β-selection cell-autonomously.
B/T cell-depleted BM from CD45.1 Itpkb+/+ and CD45.2 Itpkb-/- mice was mixed at a 1:1 ratio and injected into CD45.1/CD45.2 lethally irradiated hosts. 7 weeks later, thymocytes were analyzed by FACS. (A) Top, thymocyte expression of CD45.1 and CD45.2. The other panels show expression of the indicated markers on CD45.1+CD45.2-Itpkb+/+ or CD45.1-CD45.2+Itpkb-/- donor-derived thymocytes, using the gating strategy in Figure 2A. Numbers denote % cells per gate. (B) Chimerism of the indicated thymocyte subpopulations, expressed as mean ± SEM ratio of CD45.1-CD45.2+Itpkb-/- to CD45.1+CD45.2-Itpkb+/+ donor-derived thymocytes. (C) Mean ± SEM ratio of total DN3 cell numbers to TCRβ- DN4 cell numbers in Itpkb+/+ or Itpkb-/- donor-derived thymocytes. Significance of the indicated comparisons was analyzed as in Figure 2 (n = 3). (D) CD2 and CXCR4 expression on Itpkb+/+ (solid) and Itpkb-/- (hatched) thymocyte subsets in mixed BM chimeras. Representative of 3 independent hosts. (E) Ki67 expression in Itpkb+/+ (black) and Itpkb-/- (gray) DN3, DN3-4 and DN4 cells in mixed BM chimeras. Open histogram, Itpkb+/+ isotype staining negative control. Representative of 3 independent hosts.
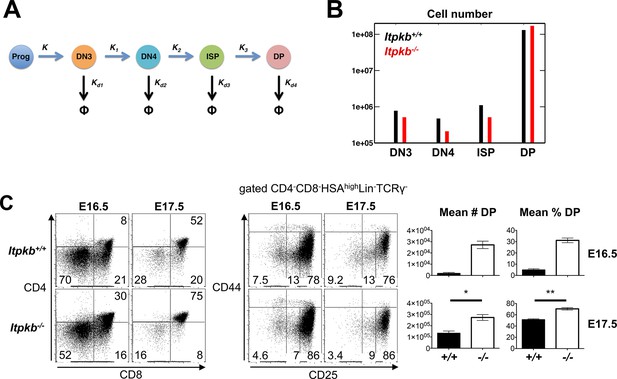
Accelerated differentiation of Itpkb-/- DN3 thymocytes.
(A) In silico analysis of β-selection kinetics in Itpkb-/- and Itpkb+/+ mice. Scheme of intrathymic DP cell development from progenitors. The velocities of relevant developmental transitions are characterized by rate constants K (identical between genotypes) and K1, K2 and K3 (development from DN3 to DP cells, set to over two-fold higher in Itpkb-/- vs. WT mice, Table 1). Rate constants Kd1-Kd4 for subset turnover via proliferation and death were considered identical between genotypes. (B) Predicted steady-state numbers of the indicated thymocyte populations in Itpkb+/+ (black) or Itpkb-/- (red) mice. (C) Left, CD4/CD8 expression on embryogenesis day (E) 16.5 or 17.5 fetal thymocytes. Center, CD44/CD25 expression on CD4-CD8-HSAhighLin-TCRγ- cells (Figure 5—figure supplement 1A). Numbers denote % cells per gate. CD4-CD8+ fetal thymocytes are ≥92% ISP (Figure 5—figure supplement 1B). Right, mean ± SEM DP cell number (#) or % in E16.5 or E17.5 Itpkb+/+ or Itpkb-/- thymi. Significance of genotype differences was analyzed as in Figure 2 (nE16.5 = 2, nE17.5 = 3). E16.5 data with t-test from another experiment in Figure 5—figure supplement 1C,D.
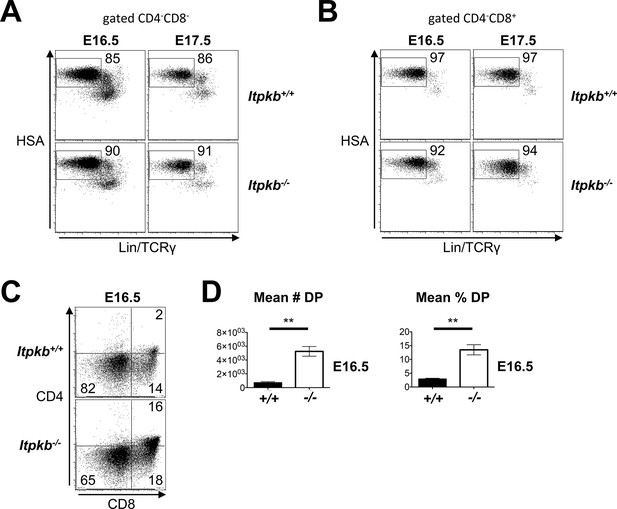
Raw and replicate data related to Figure 5C.
(A,B) HSA and combined Lin/TCRγ expression on the DN (A) and CD4-CD8+ (B) cells in Figure 5C, left panel. The HSAhighLin-TCRγ- gates in (A) were analyzed for DN cell subsets in Figure 5C, center panel. The HSAhighLin-TCRγ- gates in (B) denote ISP. ISP comprise ≥92% of CD4-CD8+ cells in E16.5 and E17.5 fetal thymi in both Itpkb+/+ and Itpkb-/- mice. (C) CD4 and CD8 expression on Itpkb+/+ or Itpkb-/- fetal thymocytes from embryos harvested on day 16.5 of embryogenesis (E16.5) from the same mother. Numbers denote % cells per gate. (D) Mean ± SEM numbers or % of DP cells in E16.5 Itpkb+/+ or Itpkb-/- fetal thymi. Significance of the indicated comparisons was analyzed as in Figure 2 (nWT = 3, nKO = 5).
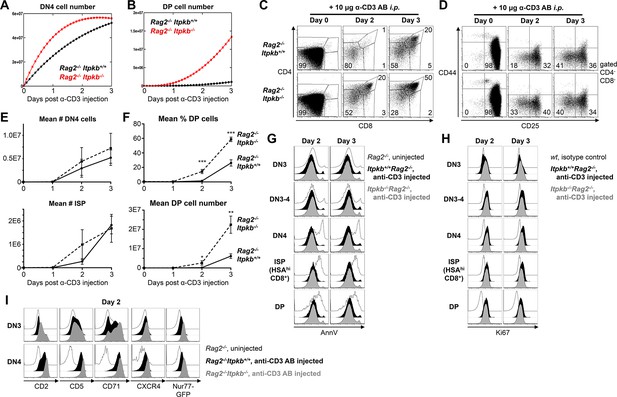
Accelerated differentiation of Itpkb-/- DN3 thymocytes.
(A,B) Mathematically predicted numbers of DN4 (A) and DP cells (B) in Rag2-/-Itpkb+/+ (black) or Rag2-/-Itpkb-/- (red) mice on the indicated days post α-CD3 antibody injection. >98% of thymocytes in Rag2-/- mice are DN3 cells (C,D), and progenitor influx within 3 days is negligible. Thus, we set K = 0 in our model for (A,B). (C,D) CD4/CD8 expression on thymocytes (C) and CD44/CD25 expression on DN cells (D) from Itpkb+/+Rag2-/- or Itpkb-/-Rag2-/- mice before (day 0), or 2 or 3 days post α-CD3 antibody injection. Gates in (D) denote DN1, DN2, DN3, DN3-4 and DN4 cells in clock-wise order, numbers % cells in the DN3 and DN4 gates. Representative of 7 independent experiments. (E) Measured mean ± SEM DN4 cell (upper panel) and ISP (lower panel) numbers in Itpkb+/+Rag2-/- (solid line) or Itpkb-/-Rag2-/- (hatched) mice before (day 0) or 1, 2 or 3 days after α-CD3 injection. Significance of the indicated comparisons was analyzed as in Figure 2. For DN4 cell numbers, n = 4, 4, 6 or 6 Rag2-/- and 4, 4, 5 or 5 Itpkb-/-Rag2-/- mice, respectively. For ISP numbers, n = 6, 5, 7 or 7 Rag2-/- and 4, 4, 6 or 5 Itpkb-/-Rag2-/- mice, respectively. (F) Mean ± SEM DP cell % (upper panel) or number (lower panel) in Itpkb+/+Rag2-/- (solid line) or Itpkb-/-Rag2-/- (hatched) mice before (day 0) or 1, 2 or 3 days after α-CD3 antibody injection. Significance of genotype differences per day was analyzed as in Figure 2. n = 6, 5, 7 or 7 Rag2-/- and 4, 4, 6 or 5 Itpkb-/-Rag2-/- mice, respectively. (G) Annexin V (AnnV) staining of DN3, DN3-4, DN4, HSAhiCD8+ ISP or DP cells from uninjected Rag2-/- (open histograms) or α-CD3 antibody injected Itpkb+/+Rag2-/- (black filled histograms) or Itpkb-/-Rag2-/-(gray filled histograms) mice two (left) or three (right) days post antibody injection. Representative of 3 independent experiments and 3–4 mice per genotype. Uninjected Rag2-/- mice contain dying cells in the DN4, CD8-ISP and DP cell gates due to failed β-selection at the DN3 stage. These serve as positive controls for the Annexin V stain. (H) Ki67 expression in DN3, DN3-4, DN4, HSAhiCD8+ ISP or DP cells from α-CD3 antibody injected Itpkb+/+Rag2-/- (black filled histograms) or Itpkb-/-Rag2-/- (gray filled histograms) mice two (left) or three (right) days post antibody injection. Representative of 2 independent experiments and 3 mice per genotype. Open histograms, day 0 WT isotype control. (I) CD2, CD5, CD71 and CXCR4 surface-levels on, and transgenic Nr4a1/Nur77-GFP expression in DN3 or DN4 thymocytes from uninjected Rag2-/- (open histograms) or α-CD3 injected Itpkb+/+Rag2-/- (black) or Itpkb-/-Rag2-/- (gray) mice 2 days post injection. The <1% CD44-CD25- negative control cells in uninjected Rag2-/- mice are non-T cells. Representative of ≥3 independent experiments.
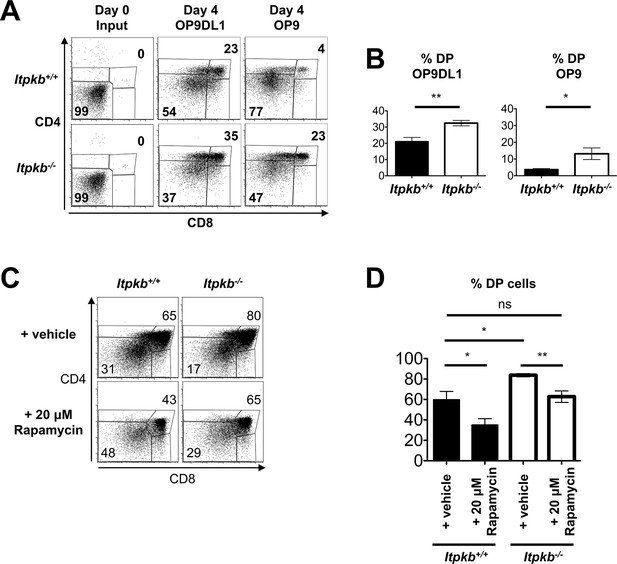
Itpkb-loss in DN3 cells causes accelerated, Notch-independent development to the DP stage.
(A,B) Sorted DN3 cells from 6.5 week old Itpkb+/+ or Itpkb-/- mice were seeded onto Delta-like 1 Notch ligand-expressing OP9DL1 or Notch ligand-free OP9 stroma cells and analyzed for CD4/CD8 expression 4 days later. (A) Representative FACS data from input (day 0) or day 4 cultures. The numbers indicate % cells in the DP or DN gates, respectively. Representative of 5 independent experiments. (B) Bar-graphs showing mean ± SEM Itpkb+/+ (black bars) or Itpkb-/- (open bars) % DP cells after 4-day culture on OP9DL1 or OP9 cells, averaged from 4 independent experiments. Significance for genotype differences was analyzed as in Figure 2 (n = 4). (C,D) Fetal thymic lobes from Itpkb+/+ or Itpkb-/- embryos harvested on day 15.5 of embryogenesis (E15.5) from the same mother were cultured in the presence of ethanol (vehicle) or 20 μM rapamycin for 4 days, harvested and analyzed. (C) Representative FACS plots of CD4/CD8 expression on total thymocytes. Numbers denote % cells in the respective gate. (D) Bar graph of mean ± SEM % DP cells for each condition and genotype from 3 independent experiments. Significance of the indicated comparisons was analyzed as in Figure 2 (n = 5).
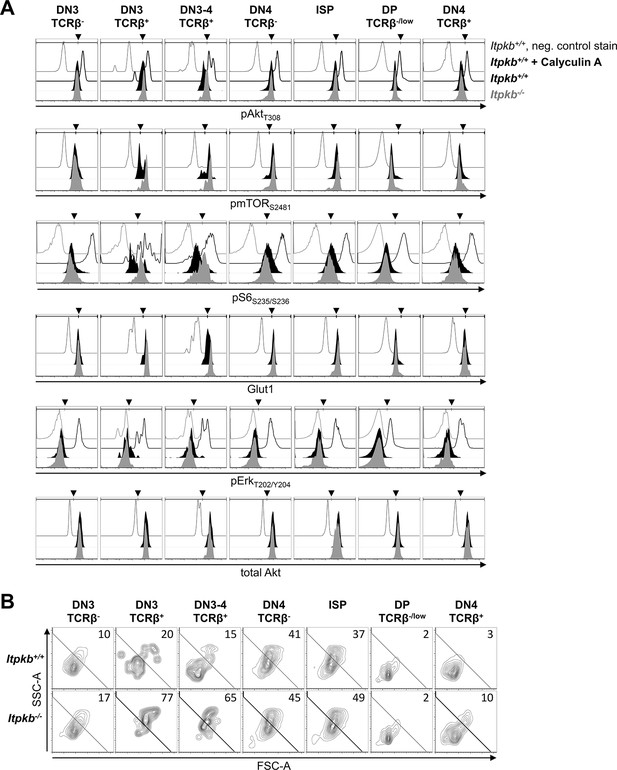
Increased pre-TCR signaling via PI3K/Akt/mTOR in Itpkb-/-DN3 cells.
(A,B) We analyzed (A) cellular content of T308-phosphorylated active Akt (pAktT308), S2481-phosphorylated mTOR (pmTORS2481), S235/S236-phosphorylated ribosomal protein S6 (pS6S235/S236), Glut1 protein, T202/Y204-phosphorylated Erk (pErkT202/Y204) and Akt protein, and (B) cell size via side/forward-scatter analysis (SSC-A/FSC-A) in the indicated thymocyte populations of Itpkb+/+ (black histograms) or Itpkb-/- (gray histograms) mice by FACS. Thin open histograms, Itpkb+/+ isotype or second antibody stained negative controls. Bold open histograms, Calyculin A-treated positive controls. Arrowheads show gate positions. In (B), numbers indicate % cells per large cell gate. Representative of at least 2 (pS6S235/S236), 3 (total Akt) or 8 (else) independent experiments.
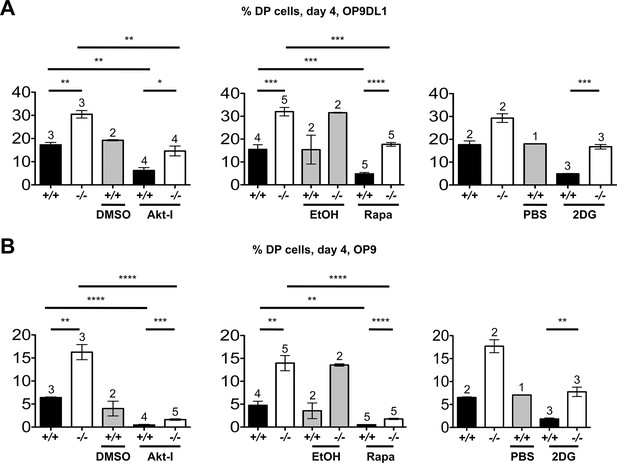
Itpkb renders β-selection Notch-dependent.
(A,B) Addition of inhibitors of Akt, mTOR or glucose metabolism reverses the accelerated development of Itpkb-/- DN3 cells and re-establishes Notch-dependence. Shown are mean ± SEM Itpkb+/+ (solid black bars) or Itpkb-/- (pen bars) % DP cells after 4-day culture on OP9DL1 (A) or OP9 (B) cells without or with addition of carrier (solid gray bars; DMSO, ethanol or PBS, respectively), 500 nM Akt-inhibitor VIII in DMSO (Akt-I, added once on day 0), 4 μM rapamycin in ethanol (Rapa, added once on day 0) or 500 μM 2-deoxy-D-glucose in PBS (2DG, added once daily), averaged from 3 (Akt-I), 4 (rapamycin) or 2 (2DG) independent experiments. Significance of the indicated comparisons was analyzed as in Figure 2. Replicate numbers are indicated above each bar. Representative FACS-data in Figure 9—figure supplement 1.
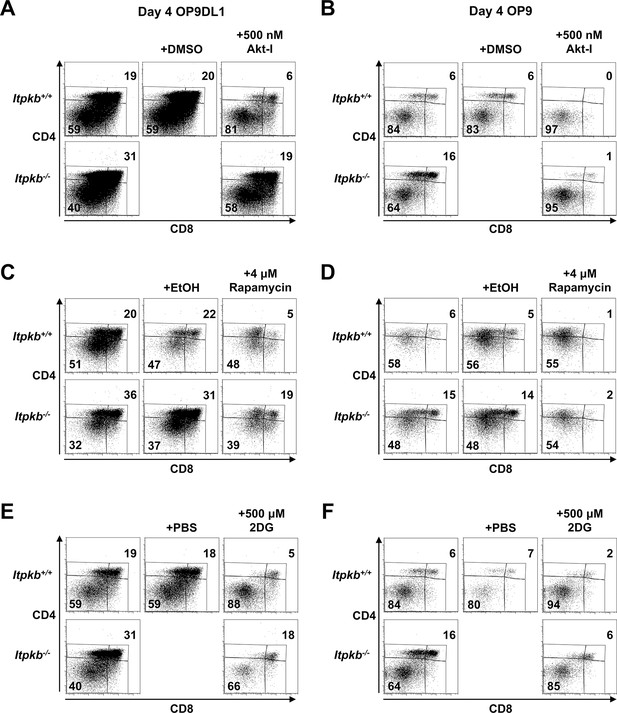
Raw FACS data from one representative experiment included in the averaged data in Figure 9.
Itpkb+/+ or Itpkb-/- sorted DN3 cells were cultured for 4 days on OP9DL1 (A,C,E) or OP9 (B,D,F) stroma cells without (left) or with once on day 0 (Akt-I, rapamycin) or once-daily (2DG) addition of carrier (center of each panel), 500 nM Akt-inhibitor VIII in DMSO (Akt-I), 4 μM rapamycin in ethanol or 500 μM 2-deoxy-D-glucose in PBS (2DG). Numbers denote % cells in the respective DP or DN gates. Representative of 3 (A,B), 4 (C,D) or 2 (E,F) independent experiments.
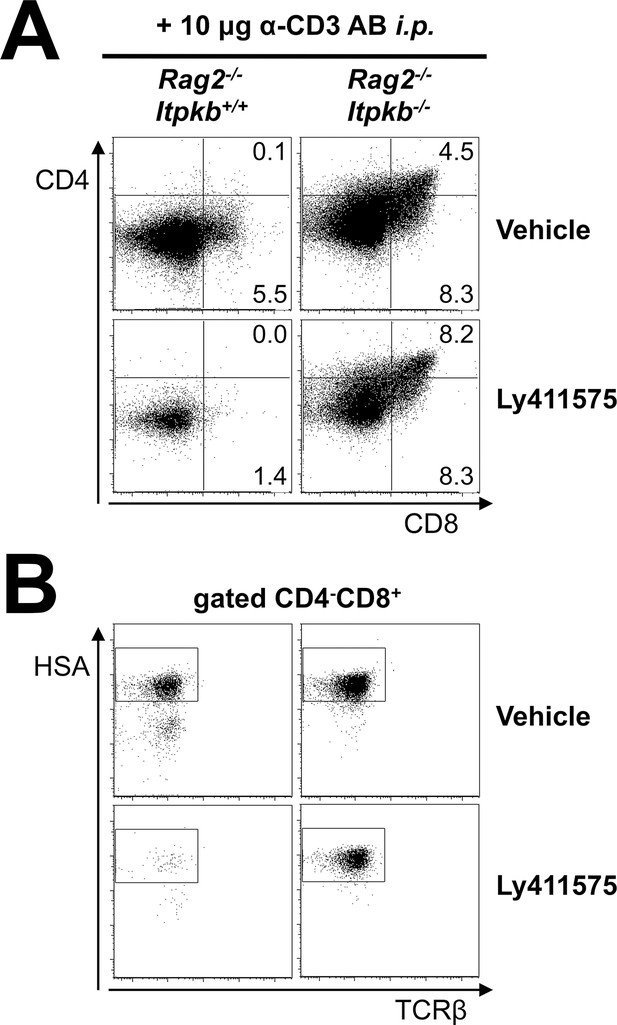
Itpkb-loss reduces the Notch-dependence of DN thymocyte development to DP cells in vivo.
Shown are (A) CD4/CD8 expression on total thymocytes and (B) HSA/TCRβ expression on CD4-CD8+ thymocytes from Rag2-/- and Rag2-/-Itpkb-/- mice two days post α-CD3 antibody injection. Starting 3–4 hr before α-CD3 injection, the mice were treated once daily with orally administered γ-secretase inhibitor LY-411,575 or vehicle (Wong et al., 2004). Numbers indicate % cells per respective gate. The gates in (B) denote CD8+HSAhigh ISP (Petrie and Zuniga-Pflucker, 2007; Xiong et al., 2011). Representative of two independent experiments (n = 3).
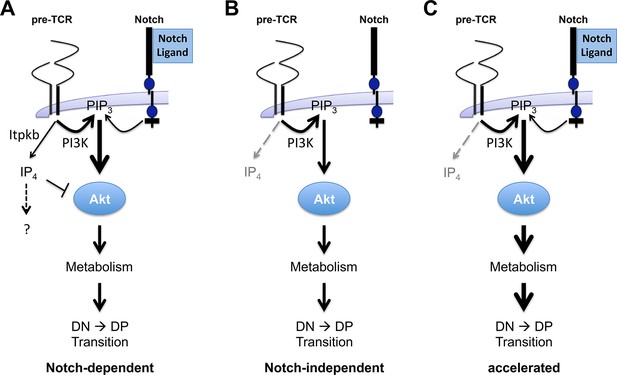
Antagonistic signaling by PI3K and Itpkb controls the kinetics and Notch-dependence of β-selection.
(A) We propose a model in which pre-TCR and Notch signaling both activate PI3K to produce PIP3 in DN3/DN3-4 cells. PIP3 then recruits and activates Akt to increase glucose metabolism via the Akt/mTOR pathway. This is required for DN3-to-DP cell differentiation. However, pre-TCR signaling also activates Itpkb to produce IP4, which competes with PIP3 for Akt PH domain binding and limits Akt recruitment, Akt and mTOR activation in pre-TCR expressing DN3/DN3-4 cells. IP4 may have additional effectors, indicated by the question mark. By limiting downstream glucose metabolism, this "IP4 brake" delays the kinetics of β-selection and renders this process dependent on Notch costimulation. (B) Without Itpkb, IP4 no more dampens Akt activation and pre-TCR signaling alone sufficiently activates Akt/mTOR signaling to trigger DP cell development in the absence of Notch engagement. (C) In the presence of Notch-signals, Akt is now hyperactivated and causes an accelerated DN3-to-DP cell differentiation.
Tables
Parameters used in our mathematical models.
| Genotype | K (cells/day) | K1 (days-1) | Kd1 (days-1) | K2 (days-1) | K3 (days-1) | Kd4 (days-1) |
|---|---|---|---|---|---|---|
| WT | 15.4 × 104 | 0.1 | 0.1 | 0.162 | 0.07 | 0.00058 |
| Itpkb-/- | 15.4 × 104 | 0.2 | 0.1 | 0.486 | 0.21 | 0.00058 |
| Rag2-/-Itpkb+/+ | 0 | 0.1 | 0.1 | 0.162 | 0.07 | 0.00058 |
| Rag2-/-Itpkb-/- | 0 | 0.2 | 0.1 | 0.486 | 0.21 | 0.00058 |





