An aspartyl protease defines a novel pathway for export of Toxoplasma proteins into the host cell
Figures
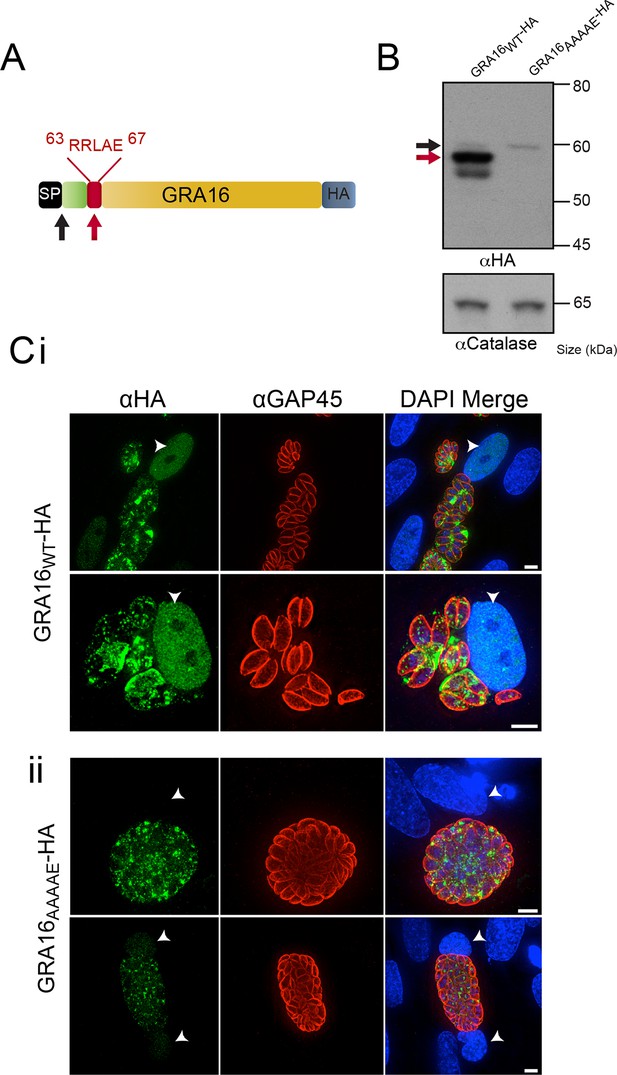
GRA16 contains a PEXEL-motif that is required for processing and export.
(A) Schematic representation of GRA16 containing an N-terminal SP for entry into the secretory pathway and a PEXEL-like (TEXEL) motif RRLAE found at residues 63–67. Arrows relate to predicted sizes of bands seen by Western blot. (B) Western blot of GRA16WT-HA and GRA16AAAAE-HA. GRA16WT-HA has three molecular weight species, the uppermost (black arrow) being consistent with SP cleaved, the middle (red arrow) consistent with TEXEL cleavage and the lowest band, which is a potential degradation product. GRA16AAAAE-HA is present as only the slowest migrating species, consistent with the expected size of signal peptide cleaved, TEXEL uncleaved. αCatalase antibodies are used as a loading control. (C) Localization of GRA16WT-HA and GRA16AAAAE-HA. (i) As previously reported, GRA16WT-HA is exported into the host cell where it accumulates in the nucleus (arrowheads) while also being present within tachyzoites and the PV space. (ii) GRA16AAAAE-HA is exported far less efficiently (a small amount can be observed in the host cell nucleus) while the majority of this protein accumulates within tachyzoites and the PV space. Scale bar is 5 μm. HA, hemagglutinin; PEXEL, Plasmodium export element; PV, parasitophorous vacuole; SP, signal peptide; TEXEL, Toxoplasma export element.
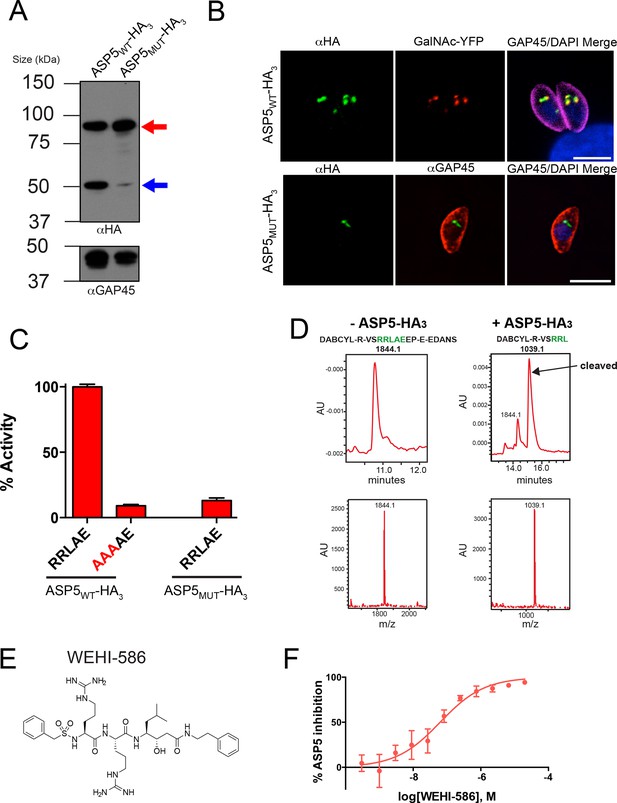
ASP5 specifically cleaves the GRA16 TEXEL.
(A) Western blot of endogenously tagged ASP5 (ASP5WT-HA3) and ectopic ASP5D431A, D682A-HA3 (ASP5MUT-HA3) in parasites shows two predominant species. The upper band (red arrow) is consistent with a signal peptidase-cleaved species and the lower (blue arrow) may be auto-activation, as it is greatly diminished for ASP5MUT-HA3. αGAP45 antibodies are used as a loading control. (B) Endogenously-expressed ASP5WT-HA3 co-localizes with the Golgi marker GalNAc-YFP (upper panel) and this localization is unaffected for the catalytic mutant enzyme (ASP5MUT-HA3) (lower panel). (C) Immunoprecipitated ASP5WT-HA3, but not ASP5MUT-HA3, cleaves GRA16 TEXEL (DABCYL-R-VSRRLAEEP-E-EDANS) but not the RRL>AAA peptide. (D) LC chromatogram (214 nm) of the fluorogenic GRA16 TEXEL peptide (upper left) incubated in buffer alone (-ASP5-HA3) with MS analysis showing the parent ion of the unprocessed fluorogenic peptide (lower left). LC chromatogram (214 nm) of the fluorogenic GRA16 TEXEL peptide after incubation at 37°C for 48 hr with ASP5 ( +ASP5WT-HA3) (upper right), showing the N-terminal product of processing within the TEXEL after leucine (DABCYL-R-VSRRL) at 15.5 min while the remaining unprocessed fluorogenic peptide is observed at 14.3 min. MS analysis showing the parent ion of the processed N-terminal cleavage product DABCYL-R-VSRRL (lower right). (E) Structure of WEHI-586 (RRLStatine). (F) Dose response curve showing inhibition of ASP5WT-HA3 activity by WEHI-586 with IC50 of 63 ± 15 nM. Data shown are mean ± standard deviation from three experiments. Scale bar is 5 μm. ASP5, Aspartyl Protease 5; HA3, triple-hemagglutinin; LC; liquid chromatography, MS; mass spectrometry.
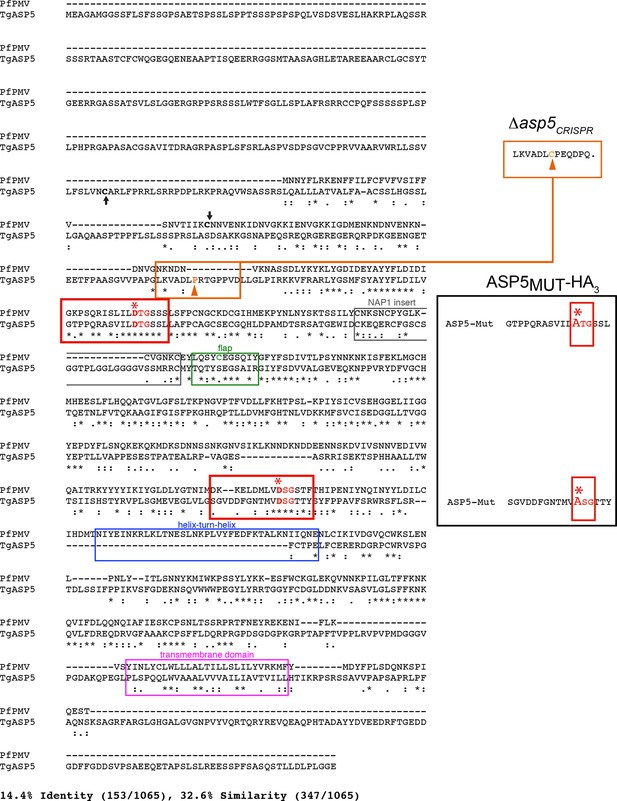
Alignment of PfPMV and TgASP5 sequences.
ClustalW alignment of the full-length predicted amino acid sequences of PfPMV (PF3D7_1323500) and TgASP5 (TGME49_242720). The alignment of the two proteins shows they share 14.4% identity and 32.6% similarity across the full-length alignment, including gaps. The predicted positions of signal peptidase cleavage are shown with black arrows, the catalytic dyads are indicated by red boxes, with ‘*’ signifying each catalytic aspartic acid residue, the ‘nepenthesin 1-type’ aspartyl protease (NAP1) fold is indicated with a gray box (NAP1 insert), the enzyme ‘flap’ that sits over the substrate binding pocket is indicated with a green box including the unique cysteine residue in PMV that is absent from ASP5, the helix-turn-helix of PMV that is absent from ASP5 is shown with a blue box, and the putative C-terminal transmembrane domain highlighted with a pink box (Hodder et al., 2015). The mutations of catalytic aspartic acid residues D>A in ASP5MUT-HA3 are shown on the right (red, D431A, D682A) and the orange arrow denotes the point of the frame shift mutation in Δasp5CRISPR parasites, which leads to a premature stop codon in the protein sequence (upper right). ASP5, Aspartyl Protease 5; PMV, plasmepsin V.
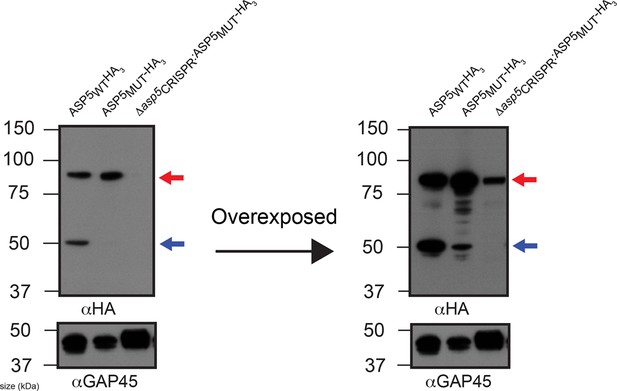
ASP5 may undergo auto proteolysis.
Immunoblot of ASP5-HA3 in (lane 1) endogenously tagged parasites, (lane 2) WT parasites ectopically expressing ASP5MUT-HA3, (lane 3) Δasp5CRISPR (right) parasites ectopically expressing ASP5MUT-HA3. A longer exposure is also shown (right panel). The ~55 kDa species is diminished in WT parasites expressing ASP5MUT-HA3 and absent in Δasp5CRISPR:ASP5MUT-HA3 parasites. The blot shown is the same as in Figure 2A, with extended panels. ASP5, Aspartyl Protease 5; HA3, triple-hemagglutinin.
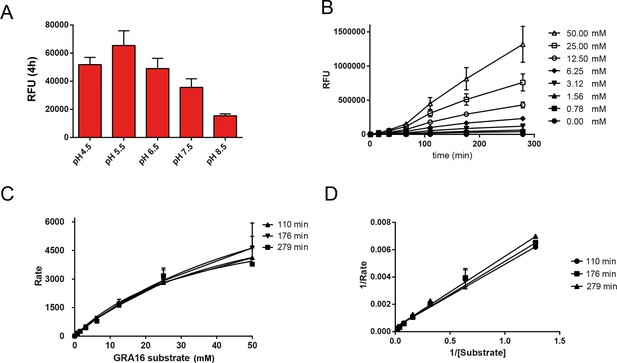
Enzymological Characterization of ASP5.
(A) ASP5WT-HA3 cleavage of a fluorogenic peptide containing the TEXEL motif from GRA16 has a pH optimum of 5.5. (B) Kinetics showing cleavage of the fluorogenic GRA16 TEXEL peptide at varying concentrations over time. (C) Michaelis–Menten curves showing the rate of cleavage (Rate = relative fluorescence units per min) of increasing concentrations of fluorogenic GRA16 TEXEL peptide by ASP5WT-HA3. The data were used to derive Km values reported in the text. (D) Burk-Lineweaver or a double reciprocal plot of the velocity of ASP5WT-HA3 as a function of the fluorogenic GRA16 TEXEL substrate concentration. Data are mean ± standard deviation of triplicate experiments. ASP5, Aspartyl Protease 5; HA3, triple-hemagglutinin; TEXEL, Toxoplasma export element.
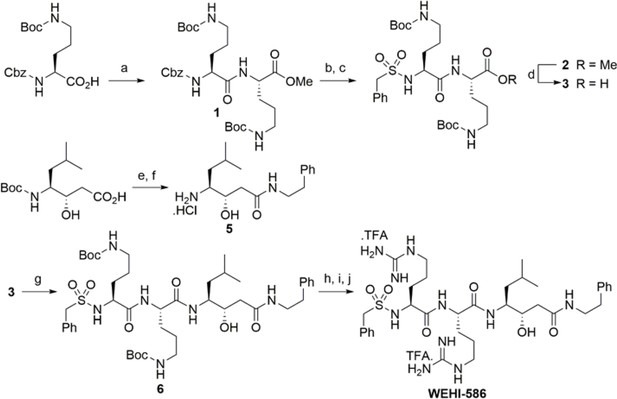
Synthesis scheme for generation of WEHI-586.
Details of WEHI-586 synthesis is outlined in Materials and methods section. Reagents and conditions: a) N,N,N′,N′-Tetramethyl-O-(1H-benzotriazol-1-yl)uronium hexafluorophosphate,O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU), Et3N, dimethylformamide (DMF), HCl.NH2-Orn(N-Boc)-OMe; b) Pd/C, H2, MeOH; c) PhCH2SO2Cl, Et3N, dichloromethane (DCM); d) LiOH.H2O, tetrahydrofuran (THF), H2O; e) HBTU, Et3N, DMF, Ph(CH2)2NH2; f) 4N HCl, dioxane; g) HBTU, Et3N, DMF, 5; h) 4N HCl, dioxane; i) Et3N, N,N'-bis-Boc-1-guanylpyrazole; j) TFA, DCM.
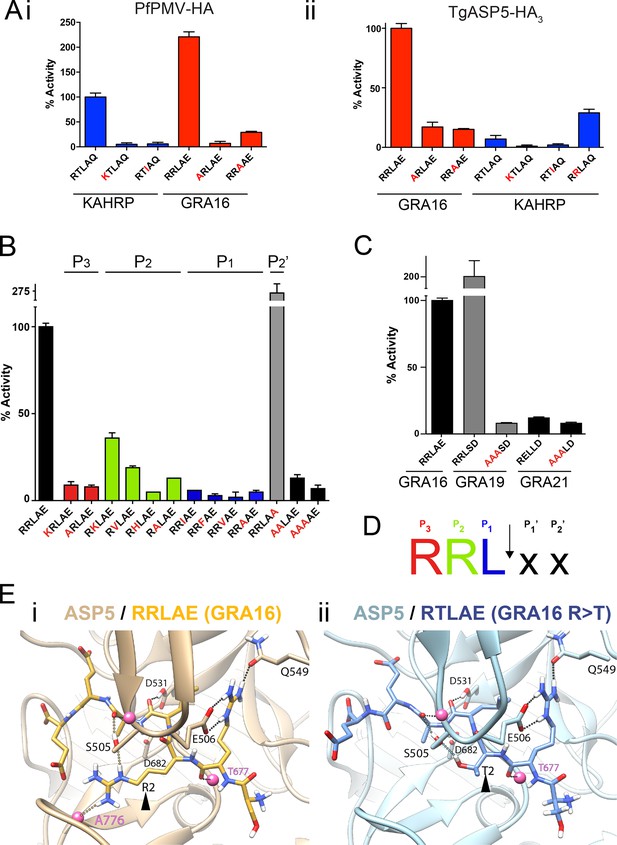
ASP5 is highly selective for ‘RRL’ substrates.
(A) (i) Activity of immunoprecipitated PfPMV-HA against KAHRP- and GRA16-based fluorogenic DABCYL/EDANS peptides. PfPMV-HA is able to cleave peptides containing KAHRP PEXEL and GRA16 TEXEL sequences but not corresponding mutants (red amino acids). Note the GRA16 ‘RRLAE’ TEXEL is cleaved approximately twice as efficiently as the KAHRP ‘RTLAQ’ PEXEL. (ii) Cleavage of substrates by immunoprecipitated TgASP5-HA3, as in (i). ASP5 cleaves the wild type GRA16 TEXEL but is unable to efficiently process the corresponding point mutants in GRA16 or any KAHRP peptides. Mutation of the P2 threonine in KAHRP for arginine (T>R) marginally increases processing. (B) Substrate specificity of ASP5-HA3 in relation to the P1, P2, P3 and P2’ positions. This protease is unable to tolerate conservative and non-conservative changes at P1, P2 or P3; however, this constriction appears to be more relaxed at P2’. (C) ASP5WT-HA3 cleaves the GRA16 TEXEL, as well as the TEXEL from the dense granule protein GRA19, but not a similar motif in GRA21, or peptides containing RRL>AAA mutations. (D) Preferred TEXEL consensus with the position of cleavage by ASP5 indicated (arrow), color-coded according to (B). (E) (i) Structural model of ASP5 in complex with the TEXEL from GRA16 (SRRLAEE) colored gold; or (ii) with a point mutant of GRA16 containing threonine at P2 (SRTLAEE) colored blue to explain why arginine is preferred at P2. Arrowheads denote the P2 position in each substrate. Heteroatoms are colored white: hydrogen, blue: nitrogen and red: oxygen. Several backbone groups in ASP5 are highlighted as pink spheres. Hydrogen bonds between the GRA16 peptides and ASP5 are shown as dotted lines; colored lines highlight the hydrogen bond interactions that differ between the two substrates. ASP5, Aspartyl Protease 5; HA, hemagglutinin; KAHRP, knob associated histidine rich protein; PEXEL, plasmodium export element; PfPMV, P. falciparum PMV; TEXEL, Toxoplasma export element.
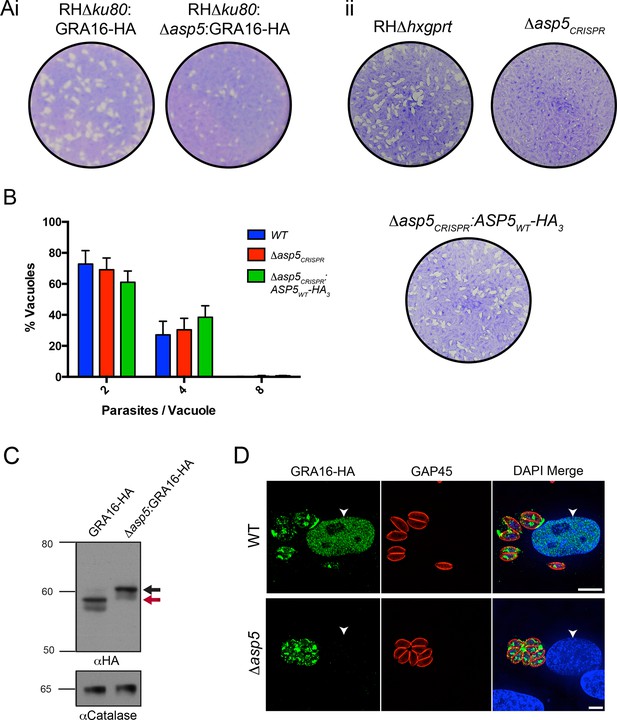
ASP5 is required for cleavage and export of GRA16.
(A) (i) A plaque assay on confluent HFF monolayers, stained with crystal violet at 7 days post infection where plaques produced by Δku80:GRA16-HA:Δasp5 parasites are smaller than those made by WT (Δku80:GRA16-HA) parasites. (ii) As in (i), where the plaques formed by Δasp5CRISPR parasites are diminished in comparison to parental wildtype (RHΔhxgprt) and Δasp5CRISPR:ASP5WT-HA3 parasites. (B) Replication assay. Tachyzoites were grown in HFFs and fixed at 16 hr post infection. Samples were stained with αGAP45 antibodies and counted. n = 3 independent experiments where > 50 vacuoles were counted, values are mean ± standard error of the mean. (C) Western blot of GRA16-HA in Δku80:GRA16-HA (lane 1) and Δku80:GRA16-HA:Δasp5 (lane 2) parasites. The black arrow corresponds to the predicted signal peptidase cleaved-species and the red arrow to the TEXEL cleaved product, as outlined in Figure 1A. Catalase antibodies are used as a loading control. (D) IFA showing GRA16-HA is exported into the host cell nucleus in otherwise WT parasites (top panel) but not in Δasp5 parasites (GFP-positive, signal diminished in comparison to the strong GRA16-HA in the 488 nm channel). White arrowheads indicate host nuclei. Scale bar is 5 μm. GFP, green fluorescent protein; HA, hemagglutinin; HFF, human foreskin fibroblasts; IFA, immunofluorescence assay; WT, wild type.
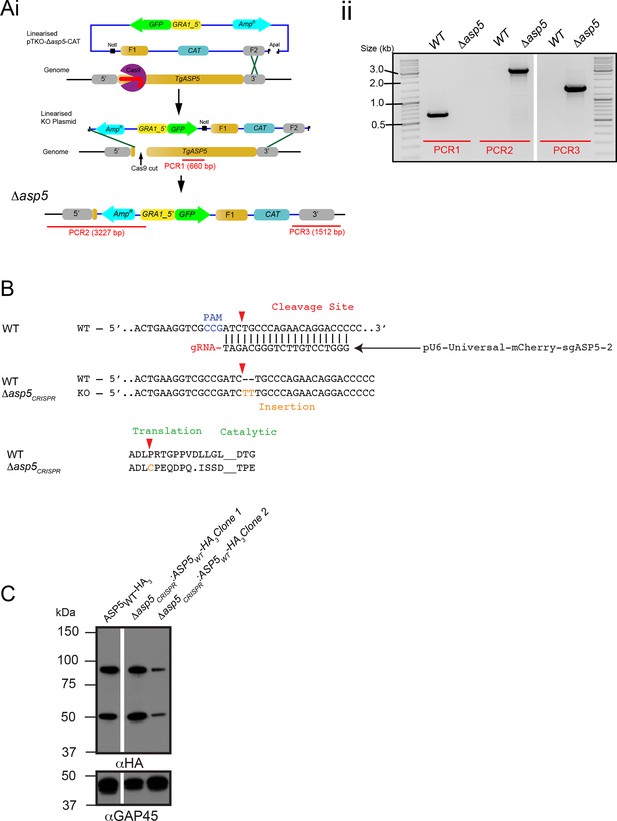
Generation and complementation of Δasp5 parasites.
(A) (i) Schematic representation of the ASP5 knockout strategy in the RHΔku80:GRA16-HA line using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 and plasmid-based recombination. (ii) PCR confirmation of a resulting Δku80Δasp5 clonal line. Sequencing of PCR products confirmed atypical integration topology (data not shown). (B) Generation of the Δasp5CRISPR parasites, where parental RHΔhx tachyzoites were transfected with pU6-Universal-mCherry-sgASP5-2 (see Materials and methods) and a clone was chosen with an insertion of ‘TT’ at the predicted Cas9 cleavage site, resulting in a frameshift mutation in the coding region of ASP5. (C) Following transfection, two clones were chosen with stably-integrated ASP5WT-HA3 in the Δasp5CRISPR line, generating the Δasp5CRISPR:ASP5WT-HA3 parasites. Δasp5CRISPR:ASP5WT-HA3 is exclusively used to refer to clone 1 in this manuscript, with the exception of Figure 7C, where both clones are used. ASP5, Aspartyl Protease 5; HA, hemagglutinin; HA3, triple-hemagglutinin; PCR, polymerase chain reaction.
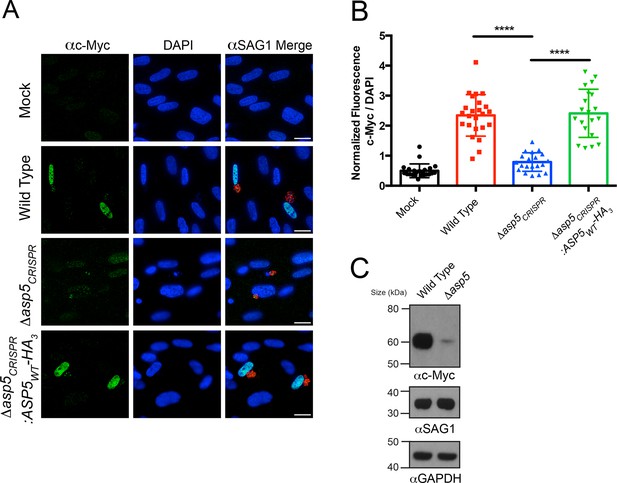
Induction of host c-Myc is ASP5-dependent.
(A) Representative IFAs 14 hr after infection of c-Myc expression in confluent HFFs. Mock-infected cells express very little c-Myc while infection with WT parasites leads to a dramatic up-regulation of this transcription factor. Δasp5CRISPR-infected cells express marginally more c-Myc than mock-infected, which is complemented by the re-introduction of ASP5 (Δasp5CRISPR:ASP5WT-HA3). (B) Quantitation of c-Myc signal (as a ratio of DAPI signal) in cells from (A), P = 0.0001, values are mean ± standard deviation, unpaired t-test, n ≥ 20 nuclei from cells infected with single vacuoles. (C) Western blot showing up-regulation of c-Myc upon wild type infection, which is drastically decreased following deletion of ASP5. αSAG1 and αGAPDH serve as parasite and host loading controls, respectively. Scale bars are 20 μm. HA3, triple-hemagglutinin; HFFs, human foreskin fibroblasts; IFA, immunofluorescence assay; WT, wild type.
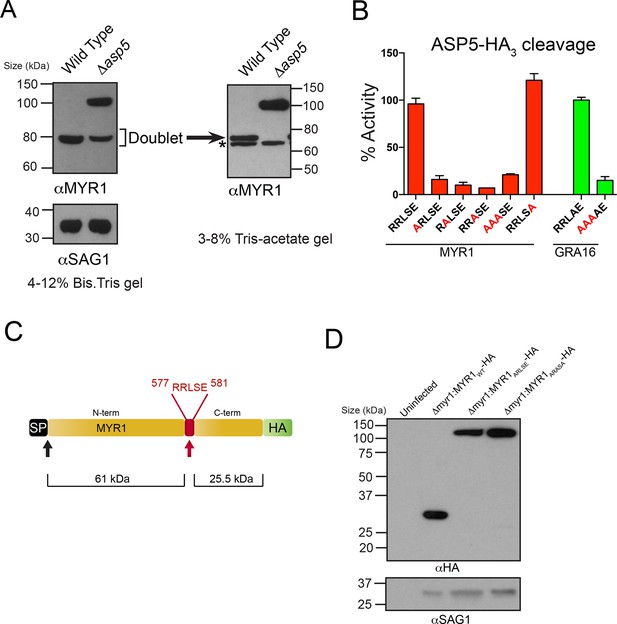
ASP5 processes the novel dense granule protein MYR1 near the C-terminus.
(A) Antibodies to the N-terminus of MYR1 show that it is processed in wild-type parasites (migrating at ~80 kDa) and a loss of processing in Δasp5 parasites resulting in the appearance of a larger molecular weight species (migrating at ~105 kDa). αSAG1 serves as the parasite loading control. For increased resolution, the samples from the left panel were separated on a 3–8% Tris-acetate gel (right panel), which revealed that the ~80 kDa band migrates at ~70 kDa on this gel and comprises a doublet, with the upper band absent in Δasp5 parasites, confirming lack of cleavage. * = suspected cross-reactive species. As a consequence of increased running time, SAG1 migrated off the 3–8% gel and was not transferred. (B) ASP5WT-HA3 cleavage of DABCYL/EDANS peptides containing the TEXEL of MYR1 and associated mutations (red residues). Peptides containing RRLSE from MYR1 and RRLAE from GRA16 are cleaved, but peptides with point mutations in P1, P2 or P3 are not cleaved. The serine at P1’ (compared to Ala in GRA16) does not interfere with cleavage by ASP5. (C) Schematic of MYR1 with an N-terminal SP, the RRLSE TEXEL at AA 557–581 and a C-terminal HA tag. (D) Immunoblot using αHA antibodies against Δmyr1:MYR1-HA parasites where Δmyr1:MYR1WT-HA runs at ~32 kDa, whereas Δmyr1:MYR1ARLSE-HA and Δmyr1:MYR1ARASA-HA mutants run at ~105 kDa. αSAG1 serves as a loading control. ASP5, Aspartyl Protease 5; HA3, triple-hemagglutinin; TEXEL, Toxoplasma export element.
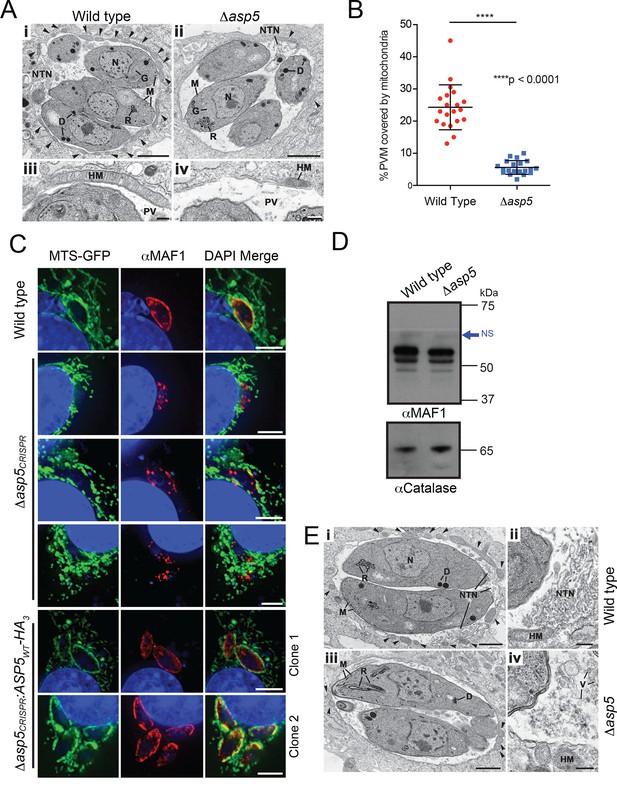
ASP5 influences efficient host mitochondrial recruitment and assembly of the NTN.
(A) Electron micrographs of intracellular WT (Δku80) (i and iii) and Δasp5 (ii and iv) tachyzoites within HFFs. Bars represent 1 µm (i,ii) and 200 nm (iii, iv). (i, ii) Low-power image showing WT (i) and Δasp5 (ii) tachyzoites containing a nucleus (N), rhoptries (R), micronemes (M), dense granules (D) and a Golgi body (G) located within a PV. Note the large number of host cell mitochondria (arrowheads) associated with the PVM and the large NTN within the PV in wild-type parasites compared to Δasp5 parasites. (iii, iv) Details from the periphery of the PV showing a large host cell mitochondrion (HM) closely applied to the PVM in the wild type (iii) compared to the smaller mitochondrion (HM) associated with the Δasp5 PV (iv). (B) Quantitation of percentage of the PVM associated with host mitochondria, 5.59 ± 2.08% for Δasp5 parasites versus 24.3 ± 6.98% for wild-type parasites, mean ± standard error of the mean, P < 0.0001, n = 20 vacuoles. (C) (i) Mouse embryonic fibroblasts expressing MTS-GFP infected for 4 hr with wild type (Δhx), Δasp5CRISPR (a non-GFP positive knock out) or two independent ASP5 complemented clones (Δasp5CRISPR:ASP5WT-HA3). Localization of MAF1 at the PVM (top panel and bottom two panels) and mislocalized in intraparasitic puncta, potentially dense granules (panels 2 and 5), are shown in red. Mitochondria (MTS-GFP) are localized at the PVM in wild-type parasites (panel 1) and Δasp5CRISPR:ASP5WT-HA3 clones 1 and 2 (panels 5–6) to a large extent, but less so in the Δasp5CRISPR parasites (panels 2–4). (ii) Immunoblot using αHA antibodies against parasites expressing ASP5WT-HA3 and complemented mutants Δasp5CRISPR:ASP5WT-HA3 clones 1 and 2 shows the parasites express similar levels of HA-tagged ASP5 (as in Figure 2A), αGAP45 serves as a loading control. (D) Western blot of MAF1 species in wild-type and Δku80Δasp5 parasites. Blue arrow shows non-specific labeling (NS), αCatalase serves as a loading control. (E) Electron micrographs of intracellular wild type (i and ii) and Δku80Δasp5 (iii and iv) tachyzoites. Bars represent 1 µm (i, iii) and 200 nm (ii, iv). (i, ii) Low-power image showing wild-type (i) and Δasp5 (iii) tachyzoites containing a nucleus (N), rhoptries (R), micronemes (M), and dense granules (D) located within the PV. The large number of host cell mitochondria (arrowheads) associated with the PVM and the large NTN within the PV in the wild type compared to the Δasp5 parasites is noteworthy. (ii) Detail of the PV of a WT parasite showing the intertwining tubules of the NTN. HM – host cell mitochondrion. (iv) Detail of the PV surrounding a Δasp5 parasite showing granular material and a few vesicles (V) but absence of the tubular network. HM – host cell mitochondrion. Scale bar is 5 μm. ASP5, Aspartyl Protease 5; GFP, green fluorescent protein; HFFs, human foreskin fibroblasts; MTS, mitochondrial targeting sequence; NTN, nanotubular network; PV, parasitophorous vacuole; PVM, parasitophorous vacuole membrane; WT, wild type.
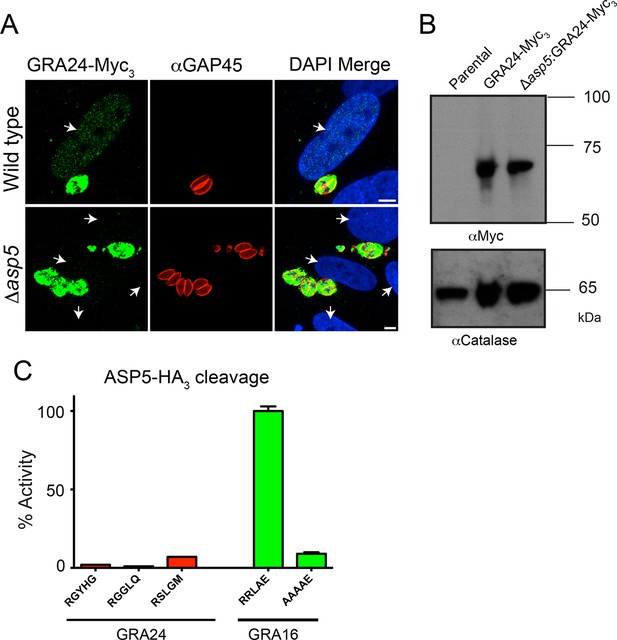
GRA24 requires ASP5 for export but not processing.
(A) Localization of GRA24-Myc3 in both WT (Δku80) and Δku80Δasp5 tachyzoites. GRA24 can be observed in the host nucleus and in the PV in WT:GRA24-Myc3 parasites, whereas this export is lost in the Δasp5:GRA24-Myc3 parasites. Arrows signify the position of host nuclei (DAPI). (B) The size of GRA24-Myc3 appears unchanged in the absence of ASP5. αCatalase serves as a loading control. (C) ASP5 cannot cleave peptides containing non-canonical TEXEL-like motifs found within GRA24, compared with cleavage of the GRA16 RRLAE peptide and AAAAE controls. Scale bar is 5 μm. ASP5, Aspartyl Protease 5; DAPI, 4',6-diamidino-2-phenylindole; PV, parasitophorous vacuole; TEXEL, Toxoplasma export element; WT, wild type.
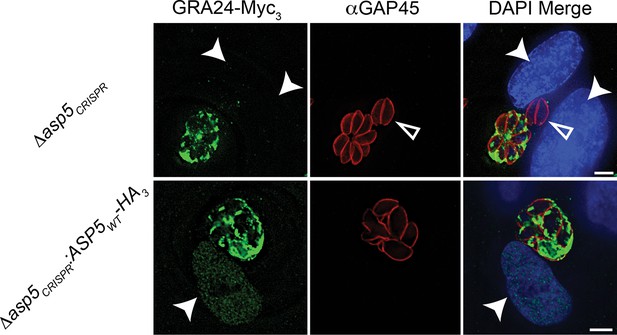
Complementation of Δasp5 parasites restores export of GRA24.
Localisation of transiently transfected GRA24-Myc3 of in both Δasp5CRISPR and Δasp5CRISPR:ASP5WT-HA3 (Clone 1) tachyzoites. Filled arrowheads signify the position of host nuclei (DAPI), open arrowheads identify non-transfected parasites and GAP45 marks the periphery of tachyzoites. Scale bar is 5 μm. DAPI, 4',6-diamidino-2-phenylindole; HA3, triple-hemagglutinin
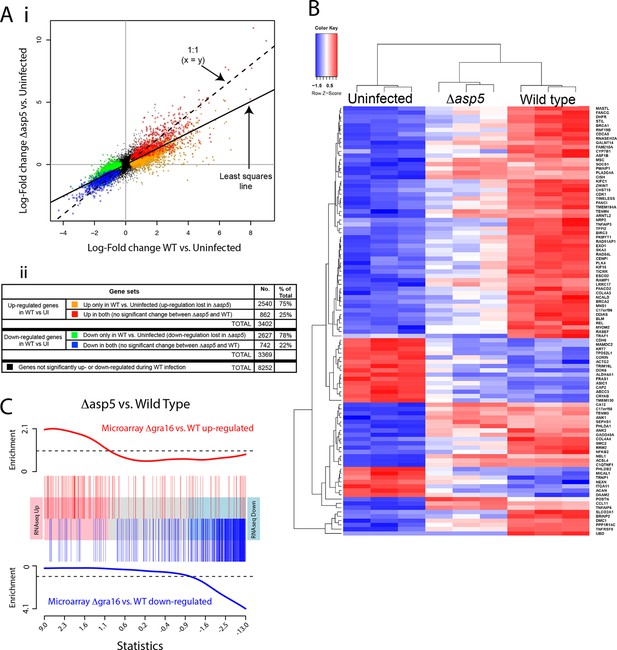
ASP5 plays a major role in changing the host cell transcriptional response induced by Toxoplasma infection.
(A) (i) Scatterplot of expression fold changes. The Y-axis shows log2-fold changes in HFFs infected with Δasp5 parasites versus uninfected HFFs (UI), while the X-axis shows log2-fold changes in HFFS infected with WT parasites (WT) vs. UI. The dashed line shows x=y. The solid line shows the least squares regression line through the origin. The regression has slope 0.6, showing that log fold changes for the Δasp5 parasites are only 60% of those for the wild-type parasites. Differentially expressed genes are color coded in the plot according to whether they change in both the WT and Δasp5 infections or only in the WT (false discovery rate < 0.05). Non-differentially expressed genes are shown in black. (ii) Numbers of genes corresponding to highlighted groups in the scatterplot. (B) Heat map of expression values for the 100 most differentially expressed genes for WT-infected HFFs versus uninfected. Z-scores are log2 counts per million, scaled to have mean 0 and standard deviation 1 for each gene. The plot shows that expression after Δasp5 infection tends to be intermediate between that of uninfected and WT-infected HFFs. (C) Barcode enrichment plot showing enrichment of Δgra16 regulated genes in the Δasp5 parasite infection expression changes. Genes are ordered from left to right in the plot from most up to most down during Δasp5 parasite infection. Specifically, genes are ranked from largest to smallest t-statistic for the Δasp5 versus WT comparison (X-axis). Genes up-regulated by Δgra16 versus WT in an independent experiment (Bougdour et al., 2013) are marked with vertical red bars. Similarly, genes down-regulated by Δgra16 in the independent experiment are marked with vertical blue bars. The worms show relative enrichment (Y-axes). The plot shows that Δasp5 up-regulated genes are strongly enriched for Δgra16 up-regulated genes (red) and Δasp5 down-regulated genes are strongly enriched for Δgra16 down-regulated genes (blue). HFFs, human foreskin fibroblasts; WT, wild type.
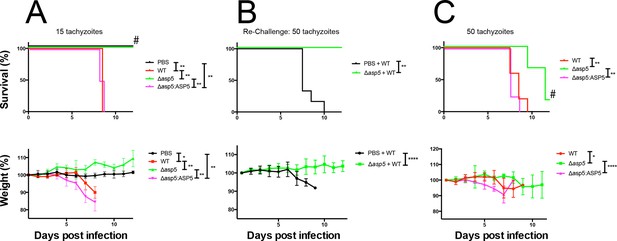
ASP5 is an important virulence factor.
(A) Four groups of six C57BL/6 mice were intraperitoneally injected with a live dose of 15 ± 3 tachyzoites or PBS alone and survival measured over a 20-day period. Mice infected with wild type (RHΔhx) and Δasp5CRISPR:ASP5WT-HA3 all succumbed to infection within 8 days, whereas all PBS-injected mice and those infected with Δasp5CRISPR parasites survived the 20 day experiment. At day 14 (#), all mice were bled and tested for antibodies against tachyzoites. Animals were weighed daily throughout the course of the experiment (lower panel) and bodyweights were compared for statistical analysis while all animals were alive. Mice injected with PBS alone maintained a stable body weight, while those infected with wild type and Δasp5CRISPR:ASP5WT-HA3 parasites lost weight beginning at day 6 and day 4, respectively, with significant weight loss evident in comparison to those injected with Dasp5CRISPR parasites by day 7. (B) At 24 days post infection, surviving mice (from A) that were injected with PBS or Δasp5CRISPR tachyzoites were re-challenged with 50 live RHΔhx parasites. The naïve PBS-injected mice all succumbed to infection by day 10, whereas those that had been injected with Δasp5CRISPR parasites were protected from death. Bodyweight was also monitored daily (lower panel) where mice previously injected with Δasp5CRISPR parasites maintained a stable bodyweight, while the naïve PBS mice began losing weight on approximately day 6. (C) A separate cohort of C57/BL6 mice was also injected with 50 live parasites to assess the effect of parasite number on virulence during infection. All mice infected with WT or and Δasp5CRISPR:ASP5WT-HA3 parasites again succumbed to infection by days 8-10, whereas there was a delay in death for the and Δasp5CRISPR-infected mice. One of these mice survived the 15-day experiment and was seropositive for antibodies against Toxoplasma (serum collected at day 14, #). Bodyweights were measured daily (lower panel). Log-rank (Mantel-Cox) testing was used to derive statistical significance for survival curves while two-way analysis of variance testing was used for bodyweight data. Values are mean ± SD. * P < 0.05, ** P < 0.005, **** P <0.0001. ASP5, Aspartyl Protease 5; WT, Wild Type.
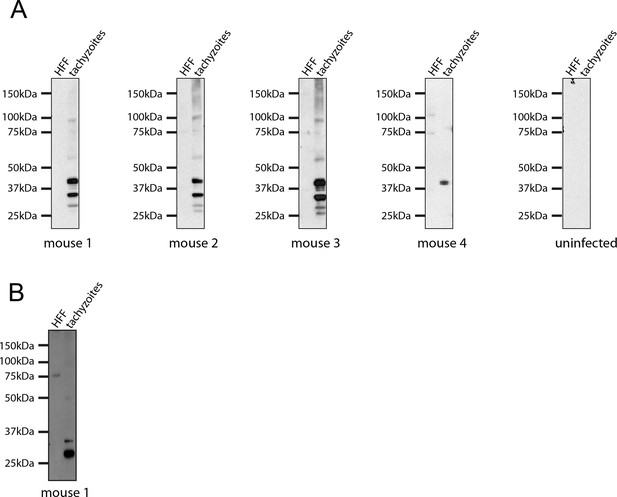
Seroconversion of Δasp5 parasites.
All mice that were alive 14 days after infection were tested for Toxoplasma sero-conversion. Toxoplasma tachyzoite and uninfected host cell lysates were probed using mouse serum at a 1:500 dilution.
Tables
Gene ontology analysis of expression changes following infection with WT and Δasp5 parasites. The four columns of the table correspond to the color-coded gene groups shown in Figure 8A. For each group of genes, the table gives the top 10 BP and MF represented in the differentially expressed genes.
Host gene expression significantly affected by loss of ASP5 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DE genes that are up-regulated in wild type versus uninfected only | DE genes that a down-regulated in wild type versus uninfected only | |||||||||||
| Biological process (BP) | GO ID | Term | Ont | N | DE | P.DE | GO ID | Term | Ont | N | DE | P.DE |
| GO:0000278 | mitotic cell cycle | BP | 884 | 284 | 2.97E-26 | GO:0006914 | autophagy | BP | 295 | 79 | 1.40E-05 | |
| GO:0090304 | nucleic acid metabolic process | BP | 3990 | 918 | 8.82E-23 | GO:0010927 | cellular component assembly involved in morphogenesis | BP | 192 | 56 | 1.85E-05 | |
| GO:0022402 | cell cycle process | BP | 1096 | 323 | 1.08E-22 | GO:0000045 | autophagic vacuole assembly | BP | 60 | 24 | 2.03E-05 | |
| GO:0007049 | cell cycle | BP | 1446 | 401 | 1.15E-22 | GO:0016559 | peroxisome fission | BP | 10 | 8 | 2.26E-05 | |
| GO:1903047 | mitotic cell cycle process | BP | 772 | 243 | 3.91E-21 | GO:0042594 | response to starvation | BP | 173 | 51 | 3.13E-05 | |
| GO:0006139 | nucleobase-containing compound metabolic process | BP | 4441 | 995 | 5.21E-21 | GO:0044782 | cilium organization | BP | 145 | 44 | 5.00E-05 | |
| GO:1901360 | organic cyclic compound metabolic process | BP | 4710 | 1040 | 5.98E-20 | GO:0007033 | vacuole organization | BP | 110 | 35 | 1.00E-04 | |
| GO:0022613 | ribonucleoprotein complex biogenesis | BP | 310 | 123 | 8.72E-20 | GO:0051146 | striated muscle cell differentiation | BP | 169 | 48 | 1.45E-04 | |
| GO:0006725 | cellular aromatic compound metabolic process | BP | 4559 | 1009 | 1.84E-19 | GO:0030031 | cell projection assembly | BP | 269 | 69 | 1.97E-04 | |
| GO:0006396 | RNA processing | BP | 532 | 180 | 2.08E-19 | GO:1903008 | organelle disassembly | BP | 162 | 46 | 1.98E-04 | |
| Molecular function (MF) | GO ID | Term | Ont | N | DE | P.DE | GO ID | Term | Ont | N | DE | P.DE |
| GO:0044822 | poly(A) RNA binding | MF | 1114 | 380 | 4.06E-42 | GO:0017049 | GTP-Rho binding | MF | 14 | 9 | 0.000104 | |
| GO:0003723 | RNA binding | MF | 1445 | 452 | 2.22E-39 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity | MF | 4 | 4 | 0.000837 | |
| GO:0003676 | nucleic acid binding | MF | 3243 | 795 | 7.02E-28 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity | MF | 5 | 4 | 0.003616 | |
| GO:1901363 | heterocyclic compound binding | MF | 4739 | 1043 | 1.67E-19 | GO:0030553 | cGMP binding | MF | 5 | 4 | 0.003616 | |
| GO:0097159 | organic cyclic compound binding | MF | 4780 | 1048 | 5.08E-19 | GO:0004499 | N,N-dimethylaniline monooxygenase activity | MF | 5 | 4 | 0.003616 | |
| GO:0003682 | chromatin binding | MF | 383 | 110 | 1.05E-07 | GO:0019905 | syntaxin binding | MF | 65 | 20 | 0.004494 | |
| GO:0043566 | structure-specific DNA binding | MF | 217 | 67 | 2.22E-06 | GO:0031697 | beta-1 adrenergic receptor binding | MF | 3 | 3 | 0.004923 | |
| GO:0017056 | structural constituent of nuclear pore | MF | 9 | 8 | 8.09E-06 | GO:0045159 | myosin II binding | MF | 3 | 3 | 0.004923 | |
| GO:0005488 | binding | MF | 10573 | 1974 | 8.11E-06 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity | MF | 3 | 3 | 0.004923 | |
| GO:0008094 | DNA-dependent ATPase activity | MF | 76 | 30 | 8.31E-06 | GO:0031210 | phosphatidylcholine binding | MF | 8 | 5 | 0.00505 | |
Host gene expression not affected by loss of ASP5 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DE genes that are up-regulated in both wild type versus uninfected and ?asp5 versus uninfected | DE genes that are down-regulated in both wild type versus uninfected and ?asp5 versus uninfected | |||||||||||
| Biological process (BP) | GO ID | Term | Ont | N | DE | P.DE | GO ID | Term | Ont | N | DE | P.DE |
| GO:0044699 | single-organism process | BP | 9400 | 689 | 1.02E-20 | GO:0003008 | system process | BP | 888 | 87 | 5.74E-10 | |
| GO:0044763 | single-organism cellular process | BP | 8542 | 642 | 5.49E-20 | GO:0032501 | multicellular organismal process | BP | 4364 | 286 | 6.37E-09 | |
| GO:0050896 | response to stimulus | BP | 5333 | 443 | 4.11E-17 | GO:0044707 | single-multicellular organism process | BP | 4228 | 277 | 1.49E-08 | |
| GO:0032501 | multicellular organismal process | BP | 4364 | 365 | 2.73E-13 | GO:0006928 | movement of cell or subcellular component | BP | 1295 | 104 | 4.60E-07 | |
| GO:0044707 | single-multicellular organism process | BP | 4228 | 354 | 8.10E-13 | GO:0045216 | cell-cell junction organization | BP | 176 | 26 | 5.96E-07 | |
| GO:0006950 | response to stress | BP | 2675 | 246 | 1.84E-12 | GO:0034330 | cell junction organization | BP | 205 | 28 | 1.13E-06 | |
| GO:0051716 | cellular response to stimulus | BP | 4476 | 367 | 4.63E-12 | GO:0048731 | system development | BP | 2914 | 195 | 1.93E-06 | |
| GO:0032502 | developmental process | BP | 3950 | 331 | 8.70E-12 | GO:0048513 | organ development | BP | 2054 | 143 | 1.01E-05 | |
| GO:0065007 | biological regulation | BP | 7817 | 570 | 2.04E-11 | GO:0034329 | cell junction assembly | BP | 182 | 24 | 1.20E-05 | |
| GO:0042221 | response to chemical | BP | 2547 | 232 | 3.00E-11 | GO:0044767 | single-organism developmental process | BP | 3888 | 243 | 1.25E-05 | |
| Molecular function (MF) | GO ID | Term | Ont | N | DE | P.DE | GO ID | Term | Ont | N | DE | P.DE |
| GO:0005125 | cytokine activity | MF | 103 | 21 | 9.63E-07 | GO:0008092 | cytoskeletal protein binding | MF | 635 | 61 | 5.31E-07 | |
| GO:0000982 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity | MF | 201 | 31 | 1.93E-06 | GO:0003779 | actin binding | MF | 299 | 36 | 7.99E-07 | |
| GO:0008009 | chemokine activity | MF | 22 | 9 | 2.92E-06 | GO:0022836 | gated channel activity | MF | 164 | 24 | 1.91E-06 | |
| GO:0005515 | protein binding | MF | 8144 | 560 | 6.08E-06 | GO:0004872 | receptor activity | MF | 592 | 53 | 2.29E-05 | |
| GO:0043565 | sequence-specific DNA binding | MF | 567 | 62 | 6.57E-06 | GO:0005216 | ion channel activity | MF | 202 | 25 | 2.40E-05 | |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | MF | 357 | 44 | 7.96E-06 | GO:0022838 | substrate-specific channel activity | MF | 204 | 25 | 2.84E-05 | |
| GO:0004857 | enzyme inhibitor activity | MF | 226 | 32 | 8.47E-06 | GO:0015267 | channel activity | MF | 217 | 25 | 7.97E-05 | |
| GO:0044212 | transcription regulatory region DNA binding | MF | 457 | 52 | 1.24E-05 | GO:0022803 | passive transmembrane transporter activity | MF | 217 | 25 | 7.97E-05 | |
| GO:0000975 | regulatory region DNA binding | MF | 459 | 52 | 1.40E-05 | GO:0038023 | signaling receptor activity | MF | 440 | 41 | 8.14E-05 | |
| GO:0001067 | regulatory region nucleic acid binding | MF | 459 | 52 | 1.40E-05 | GO:0005230 | extracellular ligand-gated ion channel activity | MF | 25 | 7 | 1.60E-04 | |
-
ASP5, Aspartyl Protease 5; BP, biological processes; DE = number of those genes that are differentially expressed genes; GO, Gene Ontology; MF, molecular functions; N = number of expressed genes annotated by the GO term; P = p-value; WT, wild type.
Additional files
-
Supplementary file 1
Primers used in this study.
- https://doi.org/10.7554/eLife.10809.021





