ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA
Figures
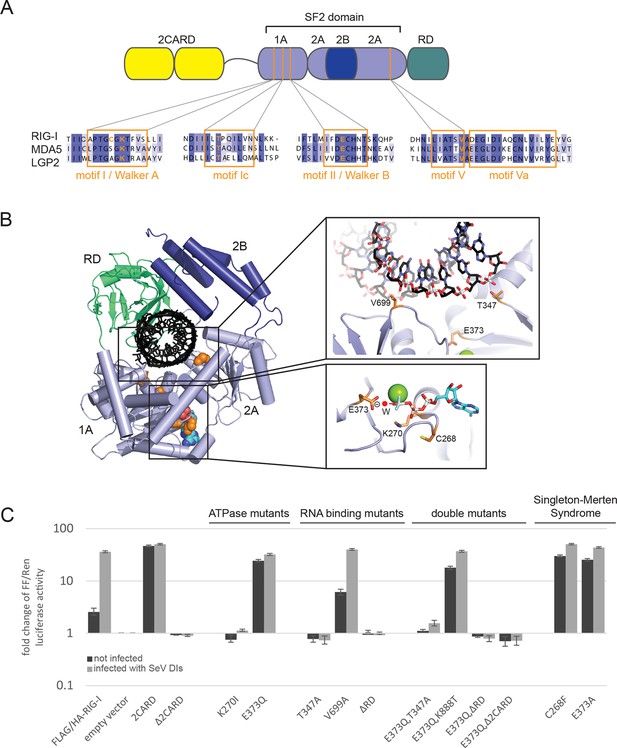
Cellular studies of RIG-I ATPase mutants in infected or non-infected cells.
(A) Location of amino acid substitutions of RIG-I SF2 domain variants used in this study (orange lines) within different RLR helicase motifs (orange squares). (B) Single amino acid substitutions (orange) within the RIG-I 3D structure (PDB: 3TMI). (C) Fold change of interferon-β (IFNβ) promoter driven luciferase activity in uninfected HEK 293T RIG-I KO cells or in cells challenged with Sendai virus defective interfering particles (SeV DIs). Cells were co-transfected with RIG-I expression vectors and p125-luc/ pCMV-RL reporter plasmids, and infected with SeV DIs 6 hr post transfection. Firefly (FF) luciferase activities were determined in respect to Renilla (Ren) luciferase activities 24 hpi. All ratios were normalized to the empty vector control. n=3–12, error bars represent mean values ± standard deviation.
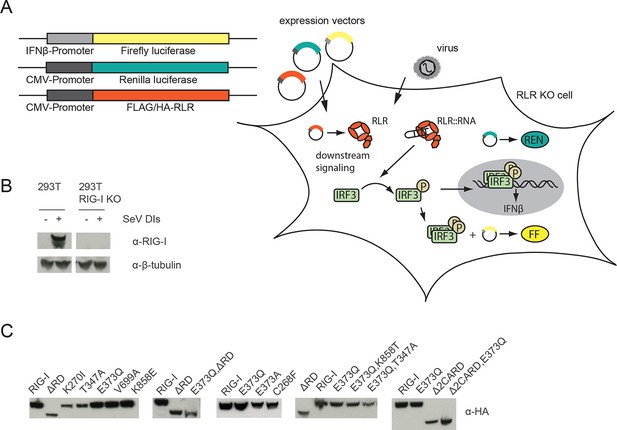
Assay for defining of the impact of RLR variant expression on RLR signaling in infected or non-infected cells.
(A) HEK 293T RIG-I KO cells were co-transfected with different expression and control vectors as indicated. RLR signaling induces an interferon-β (IFNβ) promoter driven expression of firefly luciferase (FF). Renilla luciferase (Ren) is constitutively expressed via a CMV promoter and serves as transfection control. (B) Western Blot analysis of virus-induced RIG-I expression in HEK 293T and HEK 293T RIG-I KO cells. (C) Western Blot control of overexpressed FLAG/HA tagged RIG-I variants in HEK 293T RIG-I KO cells from Figure 1, panel C.
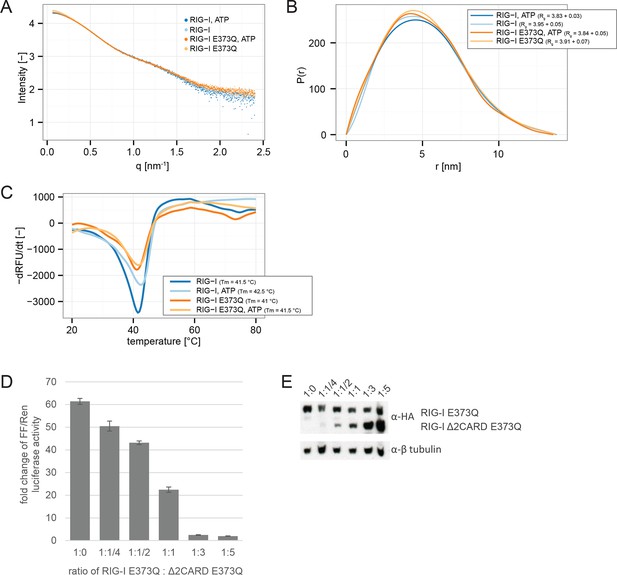
RIG-I E373Q mutation does not confer constitutive activity due to an exposed 2CARD module.
(A) Small-angle X-ray scattering (SAXS) intensity curves of RIG-I and RIG-I E373Q in presence and absence of ATP. (B) Distance distribution functions derived from SAXS data in panel A. Calculated radii of gyration (Rg) are indicated within the legend. (C) Thermal shift assays in presence and absence of ATP. Melting temperatures I are indicated within the legend. (D) Fold change of interferon-β promoter driven luciferase activity of HEK 293T RIG-I KO cells co-transfected with a RIG-I E373Q expression vector, varying concentrations of a RIG-I Δ2CARD,E373Q expression vector and p125-luc/ pCMV-RL reporter plasmids. Firefly luciferase (FF) activities were determined in respect to Renilla luciferase (Ren) activities 24 hr after transfection. All ratios were normalized to an empty vector control. n=3, error bars represent mean values ± standard deviation. (E) Control Western Blot analysis of FLAG/HA-tagged constructs from panel D.
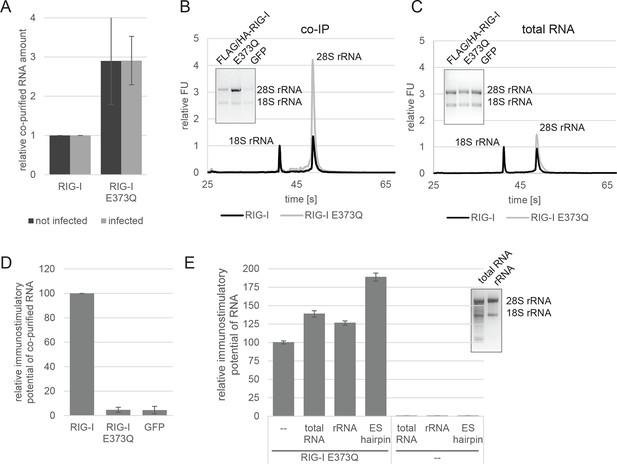
RIG-I ATP hydrolysis defective mutant E373Q recognizes the 60S ribosomal subunit in vivo.
(A) Relative RNA amount co-purified with overexpressed RIG-I or RIG-I E373Q from virus infected or non-infected HEK 293T RIG-I KO cells. n=4 (infected) or n=10 (non-infected), error bars represent mean values ± standard deviation. (B) Bioanalyzer evaluation and agarose gel separation of RNA co-purified with overexpressed RIG-I or RIG-I E373Q from non-infected HEK 293T RIG-I KO cells. Curves are normalized in respect to 18S rRNA peaks. (C) Bioanalyzer evaluation and agarose gel separation of total RNA content of non-infected HEK 293T RIG-I KO cells overexpressing RIG-I or RIG-I E373Q. Curves were normalized as in panel B. (D) Immunostimulatory potential of co-purified RNA from RIG-I, RIG-I E373Q or GFP overexpressed in measles virus (MeV), MeV-Cko-ATU-Cs or Sendai virus Cantell (SeV) infected HEK 293T RIG-I KO cells. RNA was back-transfected into HEK 293T ISRE-FF/RFP cells together with pTK-RL transfection control. Firefly luciferase (FF) activities were determined 24 hr after transfection in respect to Renilla luciferase (Ren) activity and were normalized to the immunostimulatory potential of RIG-I associated RNA. n=4, error bars represent mean values ± standard deviation. (E) Immunostimulatory potential of endogenous RNA in cells overexpressing RIG-I E373Q. RNA was co-transfected into HEK 293T RIG-I KO cells together with a RIG-I E373Q expression vector and p125-luc/ pCMV-RL reporter plasmids. FF luciferase activities were determined in respect to Ren luciferase activities 24 hr after transfection. All ratios are normalized to the RIG-I E373Q control without RNA stimulation. Purified RNA was in addition analyzed on agarose gels. n=3, error bars represent mean values ± standard deviation.
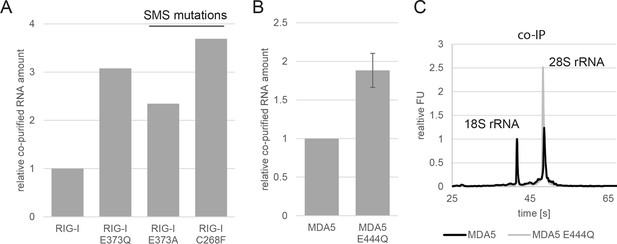
Analysis of RNA co-purified with RIG-I SMS or MDA5 variants.
(A) Relative RNA amount co-purified with overexpressed RIG-I, RIG-I E373Q or RIG-I SMS variants from non-infected HEK 293T RIG-I KO cells. (B) Relative RNA amount co-purified with overexpressed MDA5 from non-infected HEK 293T cells. n=3, error bars represent mean values ± standard deviation. (C) Bioanalyzer evaluation of RNA co-purified with overexpressed MDA5 from non-infected HEK 293T cells. Curves are normalized in respect to 18S rRNA peaks.
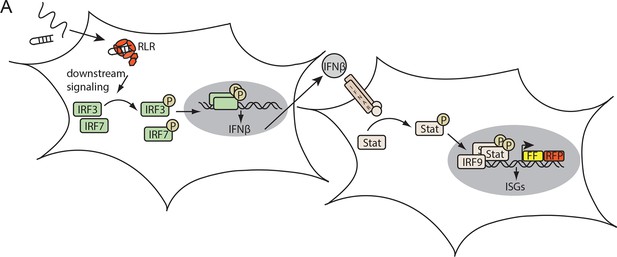
Assay for defining the immunostimulatory potential of different RNAs.
(A) Endogenous RLRs in HEK 293T ISRE-FF/RFP cells (stably express firefly luciferase (FF) and RFP under control of an interferon stimulated response element (ISRE) promoter) induce a downstream signaling cascade upon binding to transfected RNA. Subsequent interferon (IFN) expression results in activation of the STAT signaling pathway which in return induces ISRE promoter driven expression of FF luciferase.
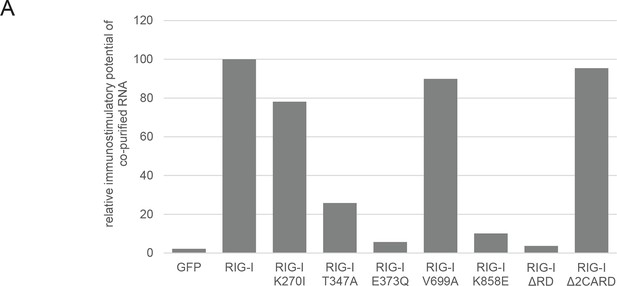
Immunostimulatory potential of co-purified RNA from Sendai virus Cantell (SeV) infected cells.
(A) HEK 293T RIG-I KO cells were transfected with the indicated RIG-I mutant or GFP expression vector. RNA co-purified with the respective overexpressed protein was back-transfected into HEK 293T ISRE-FF/RFP cells (compare with Figure 2—figure supplement 2). Firefly luciferase activities were determined 24 h after transfection and normalized to the RIG-I sample.
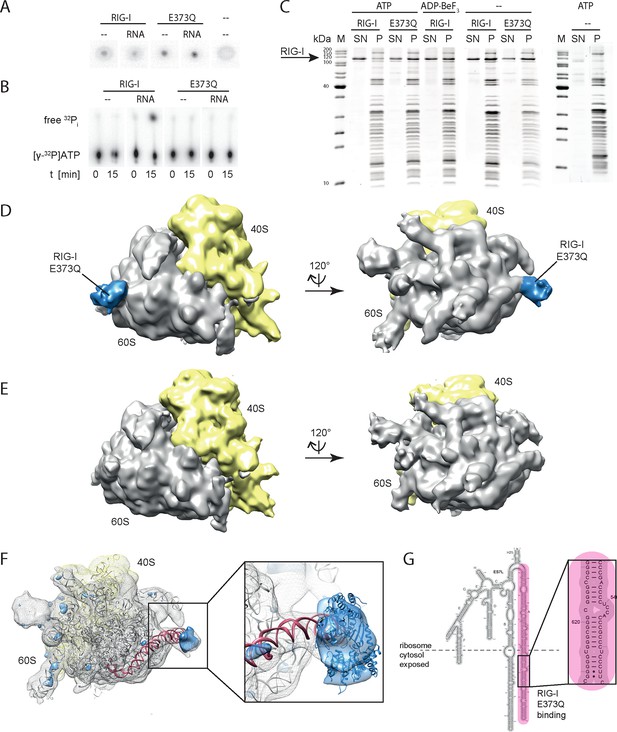
RIG-I ATP hydrolysis defective mutant E373Q recognizes the 60S ribosomal subunit in vitro.
(A) DRaCALA ATP binding assay of RIG-I or RIG-I E373Q in presence or absence of RNA. (B) ATP hydrolysis assay of RIG-I or RIG-I E373Q in presence and absence of RNA. (C) Binding studies of human 80S ribosomes with RIG-I or RIG-I E373Q in presence or absence of ATP or ADP·BeF3. Pre-formed complexes were separated on sucrose cushions via ultracentrifugation and pellet (P) as well as supernatant (SN) fractions were analyzed by SDS-PAGE. (D) Side views of a cryo-EM reconstruction of RIG-I E373Q (blue) bound to the human 80S ribosome (yellow: 40S subunit, gray: 60S subunit). Data was low pass-filtered at 15 Å. (E) Side views of a cryo-EM reconstruction of the human 80S ribosome without prior RIG-I E373Q incubation. Data filtering and color coding as in panel D. (F) Statistical difference map (left, σ = 2) of cryo-EM reconstructions in panels D and E reveals a significant additional density at expansion segment 7L A (ES7L-A, pink) into which RIG-I (PDB 3TMI) can be fitted (right, σ = 1.51). (G) Secondary structure map of the 28S rRNA ES7L (derived from (Anger et al., 2013) and zoom into RIG-I E373Q binding area. ES7L-A is indicated in pink (as in panel F).
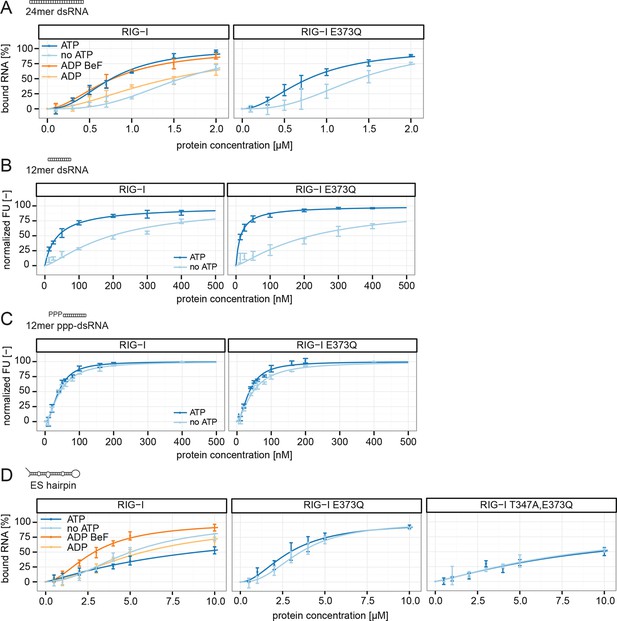
RIG-I’s ATP hydrolysis enhances RNA end recognition and removes RIG-I from RNA stems.
(A) Quantification of electrophoretic mobility shift assays of RIG-I or RIG-I E373Q incubated with 24mer dsRNA in presence or absence of ATP, ADP or ADP·BeF3 (compare with Figure 4—Figure supplement 1B). (B) Fluorescence anisotropy changes measured by titrating RIG-I or RIG-I E373Q in presence or absence of ATP into solutions containing fluorescently labeled 12mer dsRNA. (C) Fluorescence anisotropy changes measured by titrating RIG-I or RIG-I E373Q in presence or absence of ATP into solutions containing fluorescently labeled 12mer ppp-dsRNA. (D) Quantification of electrophoretic mobility shift assays of RIG-I, RIG-I E373Q or RIG-I T347A, E373Q incubated with an RNA hairpin derived from helix A of the human ribosome expansion segment 7L (ES hairpin) in presence or absence of ATP, ADP or ADP·BeF3 (compare with Figure 4—Figure supplement 1C). All binding curves were fitted using the LL.2 function of the R drc package (Cedergreen et al., 2005). n=3-6, error bars represent mean values ± standard deviation.
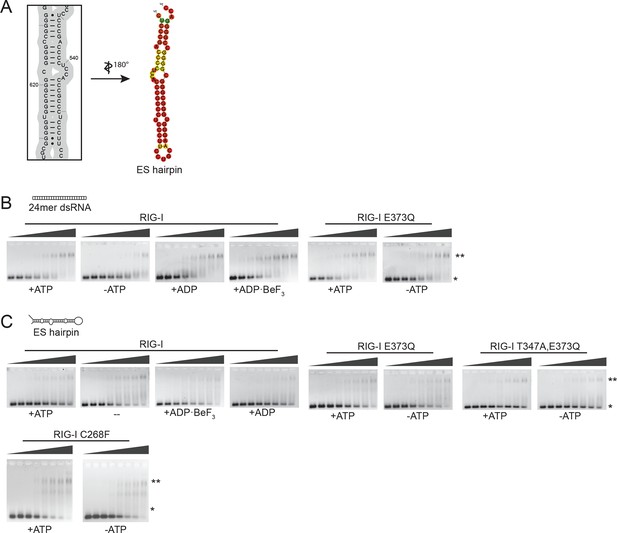
Design of the ribosomal expansion segment derived hairpin RNA, EMSA raw figures and control experiments with RIG-I C268F SMS mutant.
(A) RIG-I E373Q binding site at ES7L-A was used to design a 60b hairpin RNA (ES hairpin). RNA secondary structure was determined with the RNAfold webserver (Gruber et al., 2008). (B) Electrophoretic mobility shift assays of RIG-I or RIG-I E373Q incubated with 24mer dsRNA. Complexes were pre-formed at 37 °C for 20 min, separated on agarose gels and stained with GelRed. Free RNA bands were quantified using ImageJ. Protein concentrations (from left to right): 0, 0.1 µM, 0.3 µM, 0.5 µM, 0.7 µM, 1 µM, 1.5 µM and 2 µM. *: unbound RNA, **: protein:RNA complexes. (C) Electrophoretic mobility shift assays of RIG-I, RIG-I E373Q or RIG-I C268F incubated with ES hairpin RNA. Complexes were pre-formed, separated and stained as in panel B. Protein concentrations (from left to right): 0, 0.5 µM, 1 µM, 2 µM, 3 µM, 4 µM, 5 µM and 10 µM. *: unbound RNA, **: protein:RNA complexes.
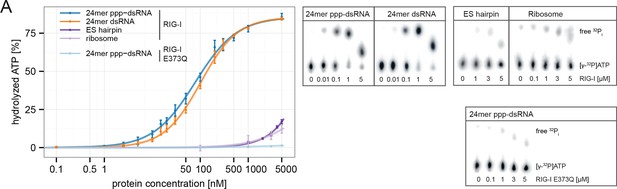
RIG-I’s ATPase activity correlates with its RNA binding affinity.
(A) Quantification of hydrolyzed [γ-32P]ATP by RIG-I or RIG-I E373Q in presence of different RNA substrates. Reactions were allowed to proceed for 20 min at 37 °C and free phosphate was separated from ATP via thin layer chromatography. Spots corresponding to labeled ATP and labeled Pi were quantified using ImageJ. All curves were fitted using the LL.2 function of the R drc package. n=3, error bars represent mean values ± standard deviation.
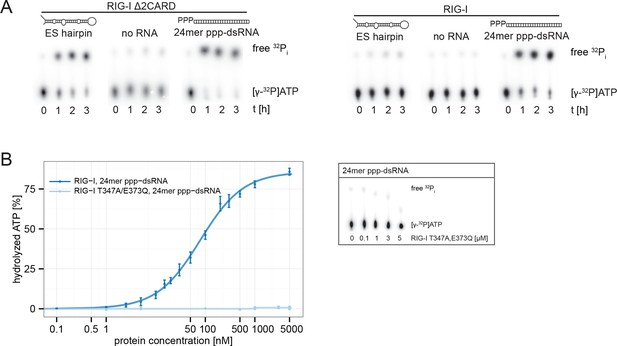
RIG-I’s 2CARD module reduces the ATP hydrolysis activity.
(A) Measurement of ES hairpin or ppp-dsRNA stimulated [γ-32P]ATP hydrolysis of RIG-I or RIG-I Δ2CARD. Reactions were monitored over 3 hr at room temperature and free phosphate was separated from ATP via thin layer chromatography. (B) Quantification of hydrolyzed [γ-32P]ATP by RIG-I T347A,E373Q in presence of 24mer ppp-dsRNA. Reactions were allowed to proceed for 20 min at 37 °C and free phosphate was separated from ATP via thin layer chromatography. Spots corresponding to labeled ATP and labeled Pi were quantified using ImageJ. Curves were fitted using the LL.2 function of the R drc package. n=3, error bars represent mean values ± standard deviation.
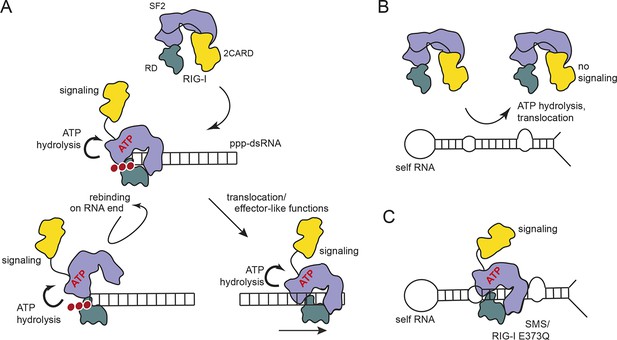
Proposed model for impact of ATP on RIG-I signaling on different RNAs.
(A) RIG-I recognizes tri- or diphosphorylated double-stranded RNA and preferentially binds to the RNA end through its regulatory domain (RD, green). Binding of ATP-SF2 (purple) to the dsRNA releases the 2CARD module (yellow) and activates the downstream signaling process. ATP hydrolysis displaces the SF2 domain from dsRNA leading to either rebinding at the RNA end (tethered by RD) or to translocation along the RNA. (B) In healthy cells, sustained binding of RIG-I to self-RNA containing dsRNA stretches is prevented by ATP hydrolysis. The SF2 domain can be sufficiently displaced because the RD does not provide a high affinity tether. (C) Mutations that allow ATP promoted binding of dsRNA and displacement of the 2CARD module, but prevent ATP hydrolysis dependent dissociation of SF2 from dsRNA, such as those underlying atypical Singleton-Merten Syndrome, will result in an unintended signaling through self-RNA.





