Extrinsic and intrinsic signals converge on the Runx1/CBFβ transcription factor for nonpeptidergic nociceptor maturation
Figures
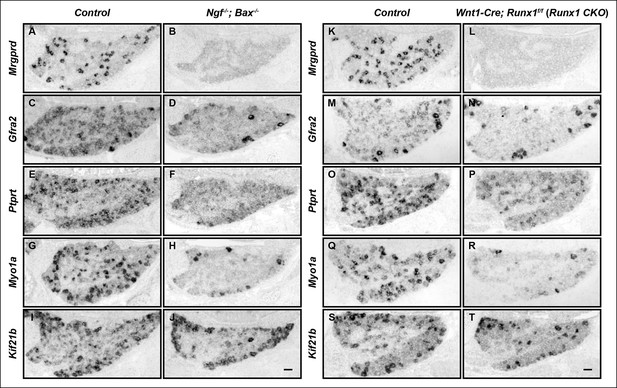
The majority of nonpeptidergic nociceptor-specific genes depend on both NGF and Runx1 for expression.
(A–J) Expression of Mrgprd (Control, 12.7% ± 2.4%; Ngf-/-Bax-/-, 0%), Gfra2 (Control, 28.7% ± 2.5%; Ngf-/-Bax-/-, 14.3% ± 1.1%), Ptprt (Control, 19.1% ± 0.3%; Ngf-/-Bax-/-, 10.3% ± 2.1%), Myo1a (Control, 26.4% ± 0.9%; Ngf-/-Bax-/-, 5.8% ± 1.5%) and Kif21b (Control, 27.2% ± 2.7%; Ngf-/-Bax-/-, 14.05% ± 1.8%) in control and Ngf-/-Bax-/- DRGs at P0, assessed by in situ hybridization. (K–T) Expression of Mrgprd (Control, 26.3% ± 1.0%; Runx1 CKO, 0.3% ± 0.2%), Gfra2 (Control, 40.0% ± 3.7%; Runx1 CKO, 9.6% ± 0.7%), Ptprt (Control, 36.0% ± 3.3%; Runx1 CKO, 10.4% ± 2.3%), Myo1a (Control, 31.1% ± 2.3%; Runx1 CKO, 5.1% ± 1.8%) and Kif21b (Control, 24.8% ± 1.7%; Runx1 CKO, 10.9% ± 0.8%) in control and Runx1 CKO DRGs at P0, assessed by in situ hybridization. Note that Ngf-/-Bax-/- and Runx1 CKO mutants display similar deficits in expression of these representative nonpeptidergic-nociceptor specific genes. For Gfra2, Myo1a, Kif21b, the apparent NGF- and Runx1-independent expression in large diameter neurons can be attributed to their expression in non-nociceptive DRG neurons. Shown are the means ± SEM for the percentage of DRG neurons expressing indicated genes based on counts from a total of at least 9 sections from three independent animals per genotype. DRG neurons were identified and counted based on combined NeuN immunostaining, which was not shown. Ngf +/-; Bax-/- or Ngf +/+ ;Bax-/- and Runx1f/f mice were used as control animals for analysis of Ngf-/-;Bax-/- and Runx1 CKO mutants, respectively. Scale bar, 50 μm. See also Figure 1—figure supplement 1.
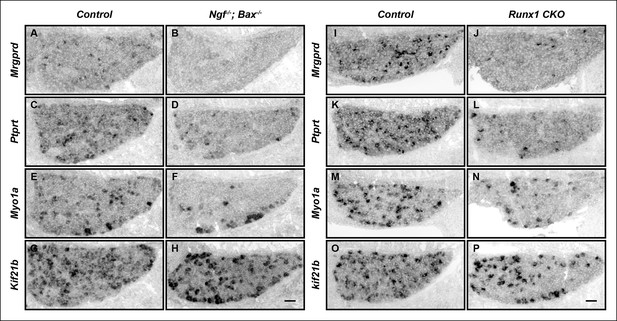
The majority of nonpeptidergic nociceptor-specific genes depend on both NGF and Runx1 for initiation of expression.
(A–P) Expression of Mrgprd, Ptprt, Myo1a and Kif21b in control and Ngf-/-Bax-/- DRGs (A-H), or in control and Runx1 CKO DRGs (I–P) at E16.5, assessed by in situ hybridization analysis. Note that expression of each of these nonpeptidergic-specific genes is impaired in both Ngf-/-Bax-/-and Runx1 CKO DRGs as early as E16.5, a time when expression is first detected, indicating a requirement of both NGF and Runx1for initiation of expression. Shown are results representative of at least two independent animals per genotype. Scale bar, 50 μm.
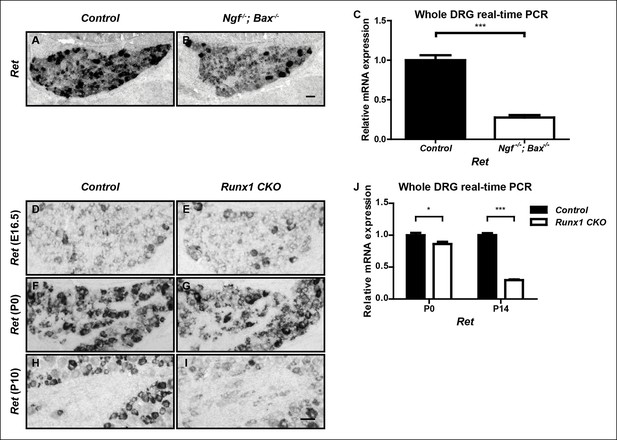
Ret is an unusual nonpeptidergic nociceptor-specific gene whose expression is differentially dependent on NGF and Runx1.
(A and B) Greatly diminished Ret expression in Ngf-/-Bax-/- DRGs compared to controls at P0, assessed by in situ hybridization. The NGF-independent Ret+ neurons are mechanoreceptors (Aβ RA-LTMRs). (C) Real-time PCR analysis of Ret expression in control and Ngf-/-Bax-/- DRGs at P0 confirms its strong NGF dependence in small diameter neurons. Statistical analysis was done using an unpaired t test, N = 4, ***p ≤ 0.001. (D–I) Ret expression in control and Runx1 CKO DRGs at E16.5 (D and E), P0 (F and G) and P10 (H and I), assessed by in situ hybridization. Note that while Ret expression is almost completely eliminated in Ngf-/-Bax-/- DRGs at P0, its expression at this same time point in Runx1 CKO DRGs is only modestly affected, indicating different temporal requirements of NGF and Runx1 for Ret expression. Shown are representative results of at least two independent animals per genotype at each time point. (J) Real-time PCR analysis of Ret expression in control and Runx1 CKO DRGs at P0 and P14 confirms the progressive nature of the Ret expression deficit in Runx1 CKO DRGs. Statistical analyses were done using unpaired t tests, N = 3 for each time point, *p ≤ 0.05, ***p ≤ 0.001. Ngf +/-; Bax-/- or Ngf +/+; Bax-/- and Runx1f/f mice were used as control animals for analysis of Ngf-/-; Bax-/- and Runx1 CKO mutants, respectively. Scale bar, 50 μm.
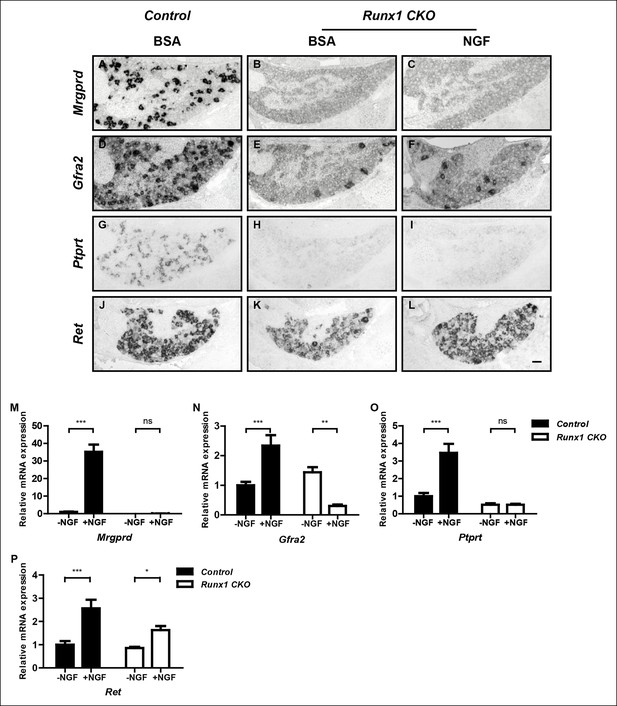
Runx1 functions downstream of NGF to mediate expression of the majority of nonpeptidergic nociceptor-specific genes, whereas it controls Ret expression at least in part by enhancing NGF signaling.
(A–L) In situ hybridization analysis of expression of Mrgprd (A–C), Gfra2 (D–F), Ptprt (G–I) and Ret (J–L) in DRGs of P2 control animals that received BSA injections, Runx1CKO animals that received BSA injections or Runx1 CKO animals that received NGF injections. Note that exogenous NGF administration fails to activate expression of nonpeptidergic-specific genes in Runx1 CKO animals, with the notable exception of Ret, suggesting that Runx1 is a downstream mediator of NGF signaling that is required for expression of the majority of nonpeptidergic-specific genes. The ability of exogenous NGF to upregulate Ret expression in Runx1 CKO animals is most consistent with an indirect role for Runx1 in regulating Ret expression, through enabling NGF signaling. Shown are results representative of at least three independent injection experiments. See also Figure 3—figure supplements 1, 2. (M–P) Real-time PCR analysis of expression of Mrgprd (M), Gfra2 (N), Ptprt (O) and Ret (P) in dissociated DRG neurons from P0 control and Runx1 CKO animals cultured in the presence or absence of NGF. Note that, with the exception of Ret, NGF-dependent expression of these nonpeptidergic-specific genes is completely abolished in the absence of Runx1, further supporting Runx1 as a downstream mediator of NGF in regulating expression of most nonpeptidergic-specific genes. Statistical analyses were done using two-way ANOVA with a Bonferroni post-test, N = 5 for M and P, N = 7 for the rest. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns non-significant. Runx1f/f mice were used as control animals for analysis of Runx1 CKO mutants. Scale bar, 50 μm.
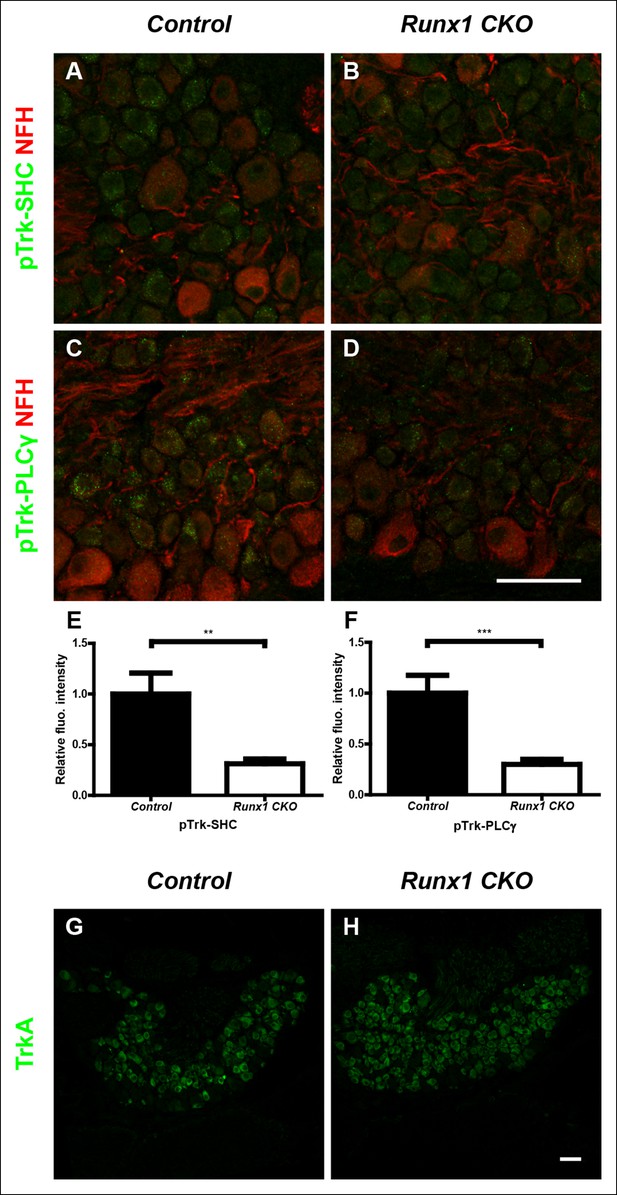
Runx1 potentiates TrkA activity without regulating TrkA expression.
(A–D) Double staining of pTrk-SHC and Neurofilament heavy chain (NFH) (A and B) or pTrk-PLCγ and NFH (C and D) in control and Runx1 CKO DRGs at P0 shows greatly diminished pTrk immunoreactivity in NFH-negative neurons in Runx1 CKO DRGs compared to controls, suggesting Runx1-dependence of NGF signaling at this time point. NFH was used to exclude myelinated TrkB, TrkC-expressing DRG neurons from analysis. (E and F) Quantification of the NGF signaling deficit based on average fluorescence intensity of pTrk-SHC or pTrk-PLCγ immunoreactivity per cell within the neuronal population that is negative for NFH. An unpaired t test was performed on data from three independent pairs of control and mutant animals, **p ≤ 0.01, ***p ≤ 0.001. (G and H) TrkA immunostaining in control and Runx1 CKO DRGs at P0 shows comparable expression in both genotypes. Shown are results representative of three independent animals per genotype. Scale bar, 50 μm.
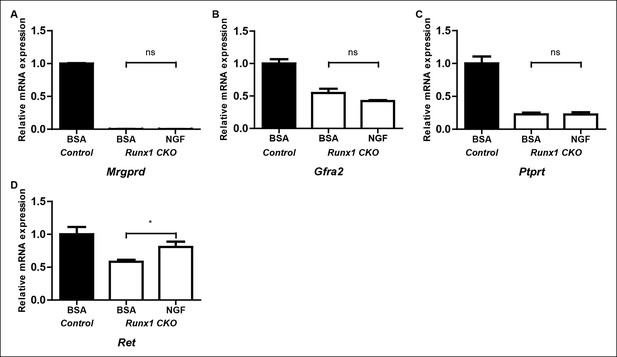
Runx1 controls expression of the majority of nonpeptidergic-specific genes independent of its stimulatory effect on NGF signaling.
(A–D) Real-time PCR analysis of expression of Mrgprd (A), Gfra2 (B), Ptprt (C) and Ret (D) in DRGs of P2 control animals that received BSA injections, Runx1CKO animals that received BSA injections or Runx1 CKO animals that received NGF injections. A one-way ANOVA with a Tukey's multiple comparison test was performed on data from three independent experiments, *p ≤ 0.05, ns non-significant.
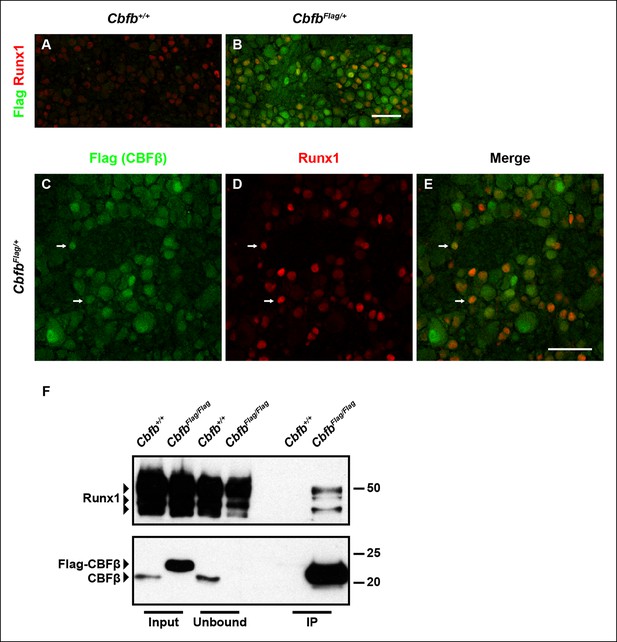
Runx1 and CBFβ are co-expressed and form a complex in DRG neurons.
(A and B) Double immunostaining of Flag and Runx1 in wildtype and CbfbFlag/+ DRGs at P0 confirms the specificity of the Flag antibody. Note that Flag immunoreactivity in wildtype DRGs is nearly undetectable. (C–E) Double immunostaining of Flag and Runx1 in CbfbFlag/+ DRGs at P0 shows extensive colocalization between Flag-CBFβ and Runx1 (arrows). Note that CBFβ is expressed in many more DRG neurons than those that are Runx1+ . See more examples in Figure 4—figure supplement 1E–G. (F) Co-immunoprecipitation experiments using a Flag antibody for immunoprecipitation from DRGs lysates from P0 wildtype and CbfbFlag/Flag animals. Subsequent detection with Runx1 and CBFβ antibodies shows that Runx1 associates with Flag-CBFβ from DRGs of CbfbFlag/Flag animals, but not wildtype controls, indicating the formation of a Runx1/CBFβ complex in the DRG. Scale bar, 50 μm.
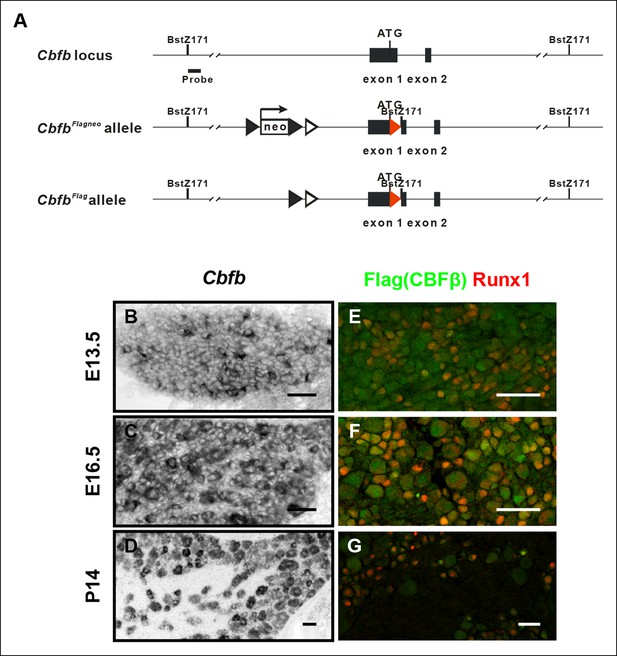
Generation of the CbfbFlag allele and more detailed characterization of the temporal and spatial patterns of Cbfb expression.
(A) Schematic showing the targeting strategy used for generation of CbfbFlagknockin mice. Following germ-line transmission, the Neo selection cassette was removed by crossing the carrier to a mouse expressing FlpE recombinase in the germ line. A Bstz171 restriction site was introduced immediately downstream of the Flag sequence to facilitate Southern screening of embryonic stem (ES) cells. Flag epitope sequences, LoxP and FRT sites are shown as red filled triangles, open and filled triangles respectively. (B–D) In situ hybridization analysis of Cbfb expression in wildtype DRGs at E13.5, E16.5 and P14 reveals a widespread pattern of expression throughout development. Note that Cbfb is expressed at varying levels in DRG neurons. (E–G) Double immunostaining of Flag (green) and Runx1 (red) in CbfbFlag/ + DRGs at E13.5, E16.5 and P14 shows a pattern of CBFβ protein similar to its mRNA distribution.
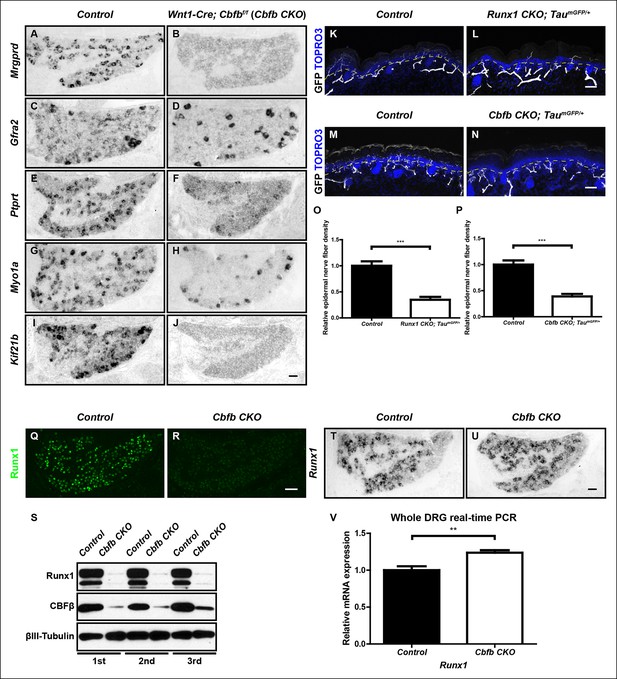
CBFβ is required for acquisition of molecular and morphological features of nonpeptidergic nociceptors.
(A–J) Expression of Mrgprd (Control, 26.9% ± 2.8%; Cbfb CKO, 0%), Gfra2 (Control, 38.8% ± 2.8%; Cbfb CKO, 11.7% ± 1.9%), Ptprt, (Control, 31.9% ± 3.2%; Cbfb CKO, 7.1% ± 2.8%), Myo1a (Control, 26.9% ± 3.2%; Cbfb CKO, 5.6% ± 0.6%) and Kif21b (Control, 20.2% ± 0.1%; Cbfb CKO, 2.4% ± 0.5%) in control and Cbfb CKO DRGs at P0 by in situ hybridization analysis. The gene expression deficits in Cbfb CKO animals phenocopy those observed in Runx1 CKO animals except for Kif21b expression. The discrepancy likely reflects Kif21b expression in proprioceptors where it presumably depends on Runx3 and CBFβ for expression. Shown are the means ± SEMs for the percentage of neurons expressing indicated genes based on counts from a total of at least 9 sections from three independent animals per genotype. DRG neurons were identified and counted based on combined NeuN immunostaining, which was not shown. See also Figure 5—figure supplement 1D, E. (K–N) GFP immunostaining of P0 hairy skin to visualize sensory innervation of the epidermis in control and Runx1 CKO animals (K and L) or control and Cbfb CKO animals (M and N) that also carry the TaumGFP allele. The TaumGFP allele was introduced to label all Cre-expressing neurons including all sensory neurons. Note that there is a dramatic reduction in fiber density specifically in the epidermis in both Runx1 CKO and Cbfb CKO animals relative to their littermate controls. The yellow dotted line denotes the epidermal-dermal junction which was drawn based on TOPRO3 counterstain (blue). (O and P) Quantification of sensory innervation of the epidermis in control and Runx1 CKO animals (O) or control and Cbfb CKO animals (P) reveals a remarkably similar reduction in the innervation density in both mutants at P0. The innervation density is defined as the fraction of area occupied by GFP+ fibers in the epidermis. An unpaired t test was performed on data from three independent animals per genotype. ***p ≤ 0.001. (Q and R) Runx1 immunostaining of control and Cbfb CKO DRGs at P0 shows almost complete loss of Runx1 proteins in the absence of CBFβ. Shown are representative images from at least three independent experiments. (S) Immunoblot analysis of expression of Runx1 and Cbfb in control and Cbfb CKO DRGs at P0 shows dramatic loss of Runx1 proteins as a result of CBFβ depletion. βIII-Tubulin serves as a loading control. Shown are results from three independent experiments. (T and U) In situ hybridization analysis of Runx1 expression in control and Cbfb CKO DRGs at P0 shows comparable levels of Runx1 transcripts in control and mutant animals. (V) Real-time PCR analysis of Runx1 expression in control and Cbfb CKO DRGs at P0 shows increased Runx1 mRNA expression in Cbfb CKO DRGs compared to control, which likely reflects an increased ratio of nociceptors to proprioceptors (data not shown). An unpaired t test was performed on data from four independent pairs of control and mutant animals, **p ≤ 0.01. Cbfbf/f mice were used as control animals for analysis of Cbfb CKO mutants. Scale bar, 50 μm.
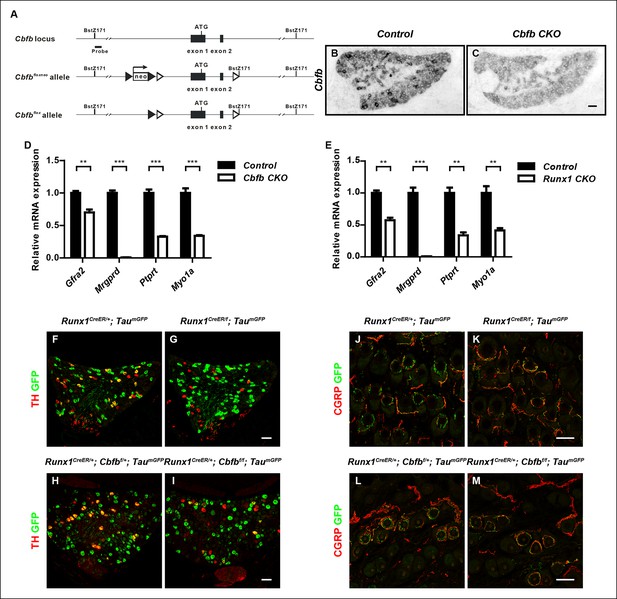
Generation of the Cbfbf allele and demonstration of a postnatal requirement for both CBFβ and Runx1 in C-LTMR development.
(A) Schematic showing the targeting strategy for generation of the Cbfbf conditional allele. Following germ-line transmission, the Neo selection cassette was removed by crossing the carrier to a mouse expressing FlpE recombinase in the germ line. A Bstz171 restriction site was introduced immediately downstream of the 3’ loxP site to facilitate southern screening of ES cells. LoxP and FRT sites are shown as open and filled triangles respectively. (B and C) Efficient gene ablation in the DRG shown by in situ hybridization analysis of Cbfb expression in control and Cbfb CKO DRGs at P0. (D and E) Real-time PCR analysis of expression of Gfra2, Mrgprd, Ptprt and Myo1a in control and Cbfb CKO DRGs (D) or control and Runx1 CKO DRGs (E) at P0 further demonstrates the similarity in nonpeptidergic-specific gene expression deficits between Cbfb CKO and Runx1 CKO mutants. Unpaired t tests were performed on data from four independent pairs of control and Cbfb CKO animals, or three independent pairs of control and Runx1 CKO animals. **p ≤ 0.01, ***p ≤ 0.001. (F-I) Expression of TH and GFP in DRGs of P21 Runx1CreER/ + ; TaumGFP/ + and Runx1CreER/f; TaumGFP/ + animals (F and G)(Control, 23.2% ± 1.6%; Runx1 mutant, 3.8% ± 2.7%) or Runx1CreER/ + ;Cbfbf/ + ; TaumGFP/ + and Runx1CreER/ + ; Cbfbf/f; TaumGFP/ + animals (H and I) (Control, 21.4% ± 1.9%; Cbfb mutant, 8.9% ± 2.0%) that received an intraperitoneal injection of tamoxifen at P2. The TaumGFP allele was introduced to indicate the neurons with active Cre expression. Note that there is a substantial reduction in the number of GFP/TH double positive neurons due to a selective loss of TH expression in the GFP + population in both Runx1 and Cbfb mutant DRGs. Shown are means ± SEM for the percentage of GFP + neurons that express TH based on counts from a total of at least 9 sections from three independent animals per genotype. (J–M) Double staining of CGRP and GFP in back hairy skin of P21 Runx1CreER/ + ; TaumGFP/ + and Runx1CreER/f; TaumGFP/ + animals (J and K) or Runx1CreER/ + ;Cbfbf/ + ; TaumGFP/ + and Runx1CreER/ + ; Cbfbf/f; TaumGFP/ + animals (L and M) that received IP injections of tamoxifen at P2. Note that in both mutant animals, there is a marked decrease in the number of GFP + longitudinal lanceolate endings characteristic of C-LTMRs, which is accompanied by an increased frequency of GFP + endings that assume a peptidergic neuron-like morphology. Shown are representative images from more than 3 independent experiments. Scale bar, 50 μm.
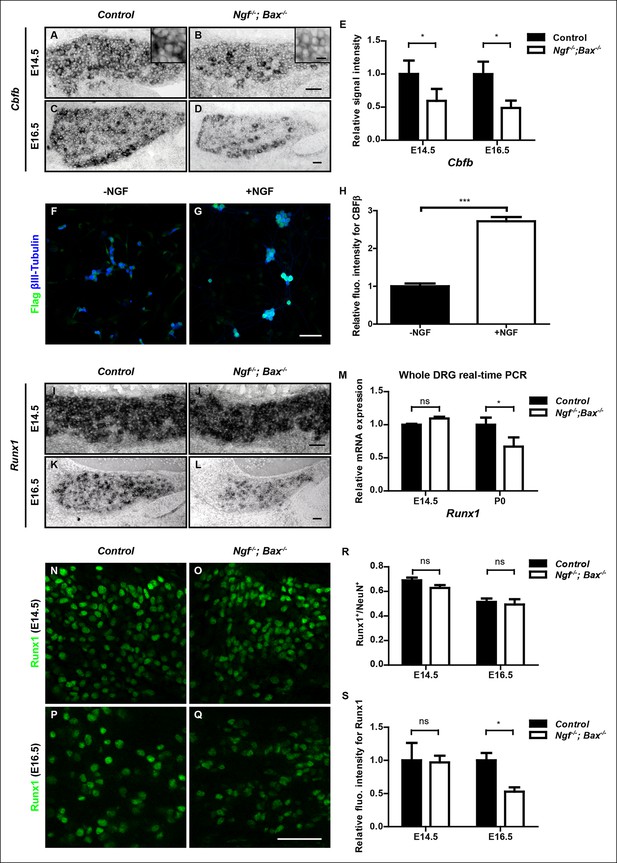
NGF regulates the Runx1/CBFβ complex through differential control of Cbfb and Runx1 expression.
(A and B) In situ hybridization analysis of Cbfb expression in control and Ngf-/-Bax-/- DRGs at E14.5 shows a significant reduction in the level of transcripts in small diameter neurons that correspond to prospective nociceptors in Ngf-/-Bax-/- DRGs compared to controls. The insets focus on nociceptor-rich regions. Scale bar for the insets, 10μm. Note that Cbfb in situ hybridization was combined with Runx3 immunostaining to exclude the Runx3+ Cbfb population from the analysis. (C and D) In situ hybridization analysis of Cbfb expression in control and Ngf-/-Bax-/- DRGs at E16.5 shows more pronounced Cbfb mRNA deficit in Ngf-/-Bax-/- DRGs. (E) Quantification of Cbfb expression deficits in nociceptors in Ngf-/-Bax-/- DRGs based on experiments described in (A–D). Intensity of in situ signal in areas devoid of Runx3+ neurons was measured. An unpaired t test was performed using data collected from 3 independent experiments for each time point. *p ≤ 0.05. See also Figure 6—figure supplement 1A. (F and G) Double staining of Flag and βIII-Tubulin in dissociated DRG neurons from P0 CbfbFlag/+ animals that were cultured without or with NGF. Note that NGF application robustly stimulates CBFβ protein expression as indicated by increased Flag immunoreactivity. (H) Quantification of the effect of NGF treatment on CBFβ protein levels based on experiments described in (F and G). CBFβ protein abundance was quantified using the average fluorescence intensity of Flag immunoreactivity per cell. An unpaired t test was performed using data collected from four independent experiments. ***p< 0.0001. See also Figure 6—figure supplement 1B–D. (I and J) In situ hybridization analysis of Runx1 expression in control and Ngf-/-Bax-/- DRGs at E14.5 shows comparable levels of Runx1 transcripts in control and mutant DRGs. Means ± SEM for relative intensity of in situ signals after normalization to the level in control DRGs is as follows: Control, 1.00 ± 0.16; Ngf-/-Bax-/-, 0.73 ± 0.12. p = 0.2079, based on an unpaired t test. (K and L) In situ hybridization analysis of Runx1 expression in control and Ngf-/-Bax-/-DRGs at E16.5 shows a reduction in the level of signal per cell in Ngf-/-Bax-/- DRGs compared to controls. Control, 1.00 ± 0.07; Ngf-/-Bax-/-, 0.49 ± 0.06. p = 0.0003, based on an unpaired t test. (M) Real-time PCR analysis of Runx1 expression in control and Ngf-/-Bax-/- DRGs at E14.5 and P0 reveals a late requirement of NGF for Runx1 expression. An unpaired t test was performed on data collected from three independent animals per genotype at each time point. *p ≤ 0.05, ns non-significant. (N–Q) Runx1 immunostaining in control and Ngf-/-Bax-/- DRGs at E14.5 (N and O) and E16.5 (P and Q) shows that the Runx1 protein deficit becomes evident in Ngf-/-Bax-/- DRGs at E16.5, which coincides with the onset of nonpeptidergic nociceptor deficits in Ngf-/-Bax-/- DRGs. (R and S) Quantification of Runx1 protein expression in control and Ngf-/-Bax-/- DRGs at E14.5 and E16.5 based on the percentage of Runx1+ neurons or the fluorescence intensity of Runx1 immunoreactivity. Note that loss of NGF specifically affects the level of Runx1 expression per cell without altering the number of Runx1+ neurons. An unpaired t test was performed on data collected from three independent animals per genotype. *p ≤ 0.05, ns non-significant. Ngf +/-; Bax-/- or Ngf +/+; Bax-/- mice were used as control animals for analysis of Ngf-/-; Bax-/- mutants. Scale bar, 50 μm.
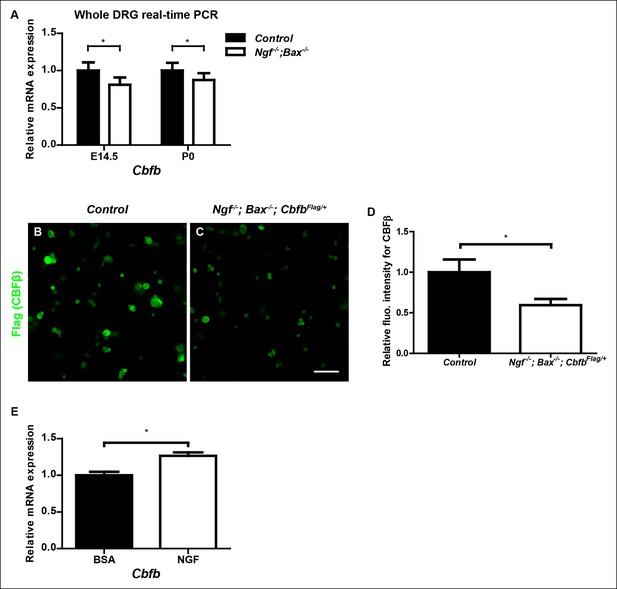
Cbfb expression is NGF-dependent in vivo.
(A) Real-time PCR analysis of Cbfb expression in control and Ngf-/-Bax-/- DRGs at E14.5 and P0 reveals early onset of NGF dependence for Cbfb expression. An unpaired t test was performed using data collected from three independent animals per genotype at each time point. *p ≤ 0.05. (B and C) Flag immunostaining of acutely dissociated DRG neurons from P0 control and Ngf-/-Bax-/- animals that are also heterozygous for CbfbFlag. (D) Quantification of NGF dependence of CBFβ protein expression based on experiments described in (A and B). CBFβ protein abundance was quantified within the βIII-Tubulin + neuronal population based on the average fluorescence intensity of Flag immunoreactivity per cell. An unpaired t test was performed using data collected from three independent experiments, *p ≤ 0.05. (E) Real-time PCR analysis of expression of Cbfb in DRGs of P2 wildtype animals that received BSA injections or NGF injections. An unpaired t test was performed on data from three independent experiments, *p ≤ 0.05. Scale bar, 50 μm.
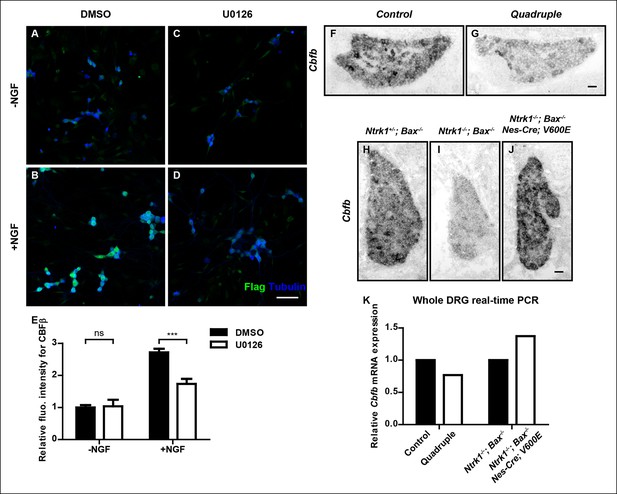
NGF promotes Cbfb expression through the ERK/MAPK signaling pathway.
(A–D) Double staining of Flag (green) and βIII-Tubulin (blue) in DMSO or U0126-treated dissociated DRG neurons from P0 CbfbFlag/+ animals that were cultured without or with NGF. U0126 is a selective inhibitor of MEK1/2, the direct activators of ERK1/2. Note that CBFβ protein expression, as defined by Flag immunoreactivity is greatly diminished in U0126-treated neurons as compared to vehicle-treated neurons, all grown in the presence of NGF. (E) Quantification of the effect of U0126 treatment on CBFβ protein levels based on experiments as described in (A–D). CBFβ protein abundance was quantified as the average fluorescence intensity of Flag immunoreactivity per cell. Statistical analysis was done using a two-way ANOVA with a Bonferroni post-test, based on data from four independent experiments. ***p ≤ 0.001, ns non-significant. See also Figure 7—figure supplement 1. (F and G) In situ hybridization analysis of Cbfb expression in control and quadruple DRGs at P0 reveals a severe deficit in Cbfb mRNA expression in DRGs when MAPK signaling is disrupted in the nervous system. A similar phenotype of varied severity was observed in 4 out of 5 mutant animals. (H–J) In situ hybridization analysis of Cbfb expression in Ntrk1 +/-; Bax-/-, Ntrk1-/-; Bax-/- and Ntrk1-/-; Bax-/-; Nes-Cre; V600E DRGs at E18.5 shows that constitutive activation of MAPK signaling leads to a dramatic increase in Cbfb expression in TrkA-deficient animals. Shown are representative images from two independent experiments. (K) Real-time PCR analysis of Cbfb expression in the same set of loss-of-function and gain-of-function mouse models as described in (F and G) and (H–J) at P0 further demonstrates the necessity and sufficiency of MAPK signaling for NGF-dependent Cbfb expression in vivo. Shown are averages from two independent experiments after normalization to littermate control. Scale bar, 50 μm.
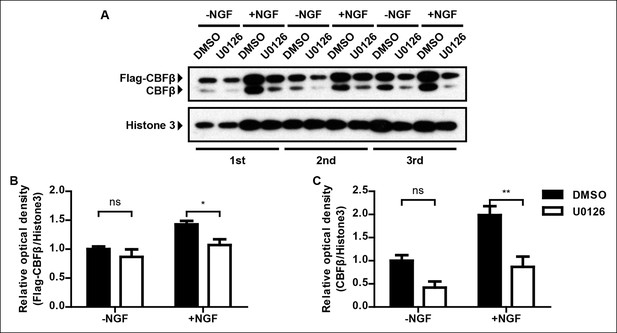
In vitro evidence for the necessity of MAPK signaling for NGF-dependent CBFβ expression.
(A) Immunoblot analysis of expression of Cbfb in DMSO or U0126-treated dissociated DRG neurons from P0 CbfbFlag/ + animals that were cultured in the presence or absence of NGF. Histone 3 serves as a protein loading control. (B and C) Quantification of the effect of U0126 on NGF dependence of Flag-CBFβ or CBFβ expression based on (A). Inactivation of MAPK signaling by U0126 attenuates NGF-dependent CBFβ expression. The levels of Flag-CBFβ or CBFβ protein were determined by densitometry, and normalized to that of Histone 3. The relative protein abundance of Flag-CBFβ or CBFβ after normalization to the level in DMSO-treated cultures that were grown without NGF was subject to a two-way ANOVA test with a Bonferroni post-test, N = 3. *p ≤ 0.05, **p ≤ 0.01, ns non-significant.
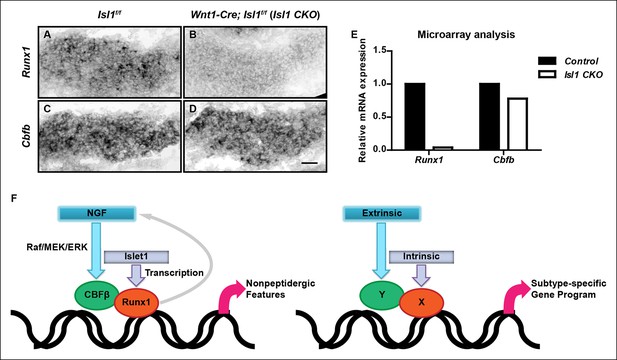
Islet1 is required for initiation of Runx1, but not Cbfb expression.
(A–D) In situ hybridization analysis of expression of Runx1 (A and B) and Cbfb (C and D) in control and Isl1 CKO DRGs at E12.5 shows that Islet1 deficiency abolishes expression of Runx1 but not Cbfb at an early age. Shown are representative images from two independent experiments. (E) Microarray analysis of E12.5 control and Isl1 CKO DRGs further confirms the differential dependence of expression of Runx1 and Cbfb on Islet1. Shown are average expression levels from two independent experiments that are normalized to the control levels for each gene. Expression levels have been normalized using globe scaling. Isl1f/f mice were used as control animals for analysis of Isl1 CKO mutants. (F) Schematics illustrating a molecular mechanism underlying specification of nonpeptidergic nociceptors and its general implication in the context of subtype specification. The extrinsic cue NGF and the intrinsic cue Islet1 coordinately regulate the Runx1/CBFβ complex, a nonpeptidergic nociceptor transcription factor complex, by preferentially targeting Cbfb and Runx1 for transcriptional regulation, respectively. Furthermore, the Runx1/CBFβ complex, through an unknown mechanism, enhances the level of NGF-TrkA signaling, resulting in a positive feedback loop between NGF-TrkA signaling and Runx1/CBFβ complex. This gene regulatory mechanism not only underscores the importance of interplay between extrinsic and intrinsic factors during multilineage differentiation, but also illustrates how such interplay can control cell-fate decisions through the convergence of extrinsic and intrinsic signals at the level of a heterodimeric, lineage-specific transcription factor complex. Scale bar, 50 μm.
Tables
Primers used for real-time PCR analysis
| Cbfb | F-TCGAGAACGAGGAGTTCTTCAGGA | R-AGGCGTTCTGGAAGCGTGTCT |
| Runx1 | F-GCAGGCAACGATGAAAACTACT | R-GCAACTTGTGGCGGATTTGTA |
| Mrgprd | F-TGCTGCTGGAAACACTTCTAGGGA | R-GCTGCTGTCAAGAGTGGAGTTCAT |
| Gfra2 | F-TCGTACAGACCACTTGTGCC | R-ATCAAACCCAATCATGCCAG |
| Ptprt | F-ACCTGCTTCAACACATCACCCAGA | R-TTCATCTTCCTTGGCTGTGTCCCA |
| Myo1a | F-ACAGGTGCTTCAACACAGCCAATC | R-GCCCTTAAACAGTTCACTGGCACA |
| Ret | F-TCAACCTTCTGAAGACAGGCCACA | R-ATGTCAGCAAACACTGGCCTCTTG |
| PGP9.5 | F-CAGACCATCGGAAACTCCTG | R-CACTTGGCTCTATCTTCGGG |
| GAPDH | F-ATGCCTGCTTCACCACCTTCTT | R-ATGTGTCCGTCGTGGATCTGA |
Additional files
-
Supplementary file 1.
Microarray analysis of genes that are differentially expressed in E16.5 DRGs of control and Runx1 CKO animals
- https://doi.org/10.7554/eLife.10874.019





