Ataxin-1 oligomers induce local spread of pathology and decreasing them by passive immunization slows Spinocerebellar ataxia type 1 phenotypes
Figures
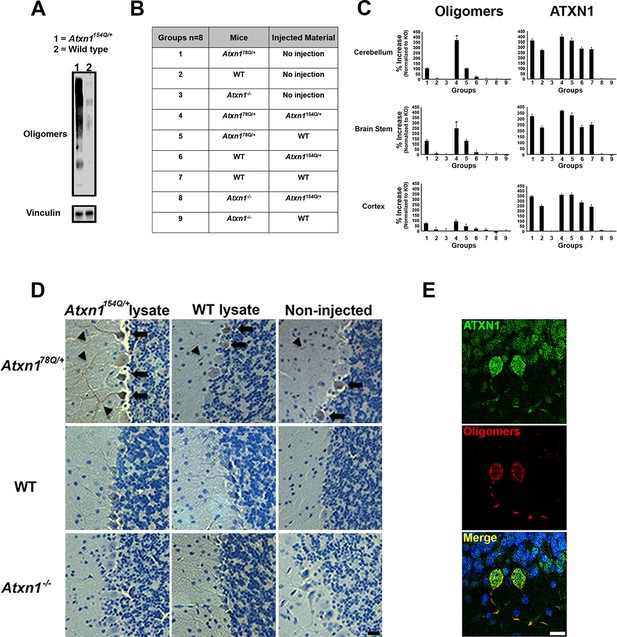
ATXN1 oligomers propagate further ATXN1 oligomerization in vivo.
(A) Representative western blot brain lysates used for in vivo injections. F11G3 was used to detect oligomers (WT and Atxn1154Q/+, cerebellar samples). (B) Table of mouse genotypes and treatments used in the in vivo propagation assay. (C) ELISA for oligomers (F11G3, left panels) and ATXN1 (11750, right panels) was performed on WT, Atxn178Q/+ and Atxn1-/- mice injected with cerebellar lysate (WT or Atxn1154Q/+) in the indicated brain regions. x axis indicated groups from (B) * denotes p<0.05, ANOVA followed by Bonferroni’s post hoc test. (D) Representative histological staining for oligomers (F11G3) in groups indicated in (B) in the cerebellum. Arrowheads indicate the accumulation of oligomers in dendrites, arrows indicate their presence in the soma of PCs. Scale bar 15 μm. (E) Double staining using anti-ATXN1 antibody (green) and anti-oligomer antibody (red) confirmed the presence of ATXN1 oligomers in Purkinje cells of Atxn178Q/+ mice injected with Atxn1154Q/+ cerebellar soluble fraction. Scale bar 15 μm.
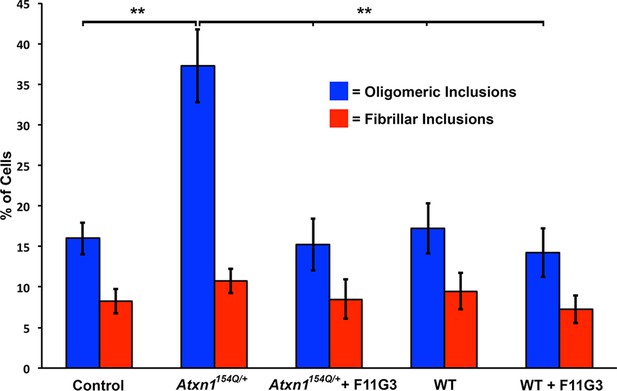
Anti-oligomer antibody F11G3 blocks the formation of oligomers in cells treated with ATXN1 oligomers from Atxn1154Q/+mouse cerebellum.
Quantification of the percentage of mRFP-ATXN1(82Q) cells with oligomeric (blue bars) or fibrillar (red bars) inclusions following treatment of Atxn1154Q/+or WT mouse cerebellum alone or pre-incubated with F11G3. ** denotes p<0.01, ANOVA followed by Bonferroni’s post hoc test.
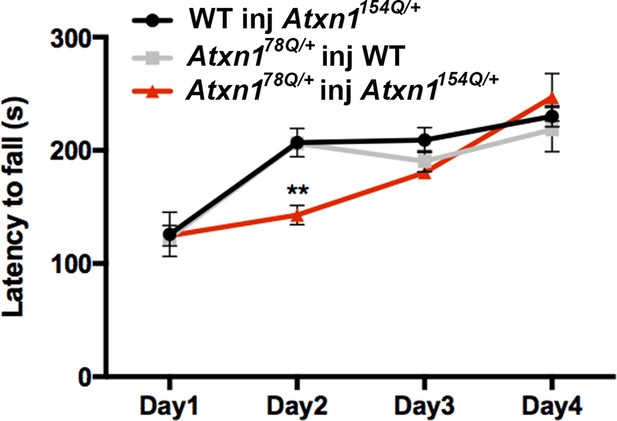
Oligomer propagation is accompanied by slight motor deficit in Atxn178Q/+ mice.
Rotarod assay over a four-day period (four trials per day, averaged), 3 months following lysate injection. n = 8 per genotype. ** denotes p<0.01, ANOVA followed by Bonferroni’s post hoc test.
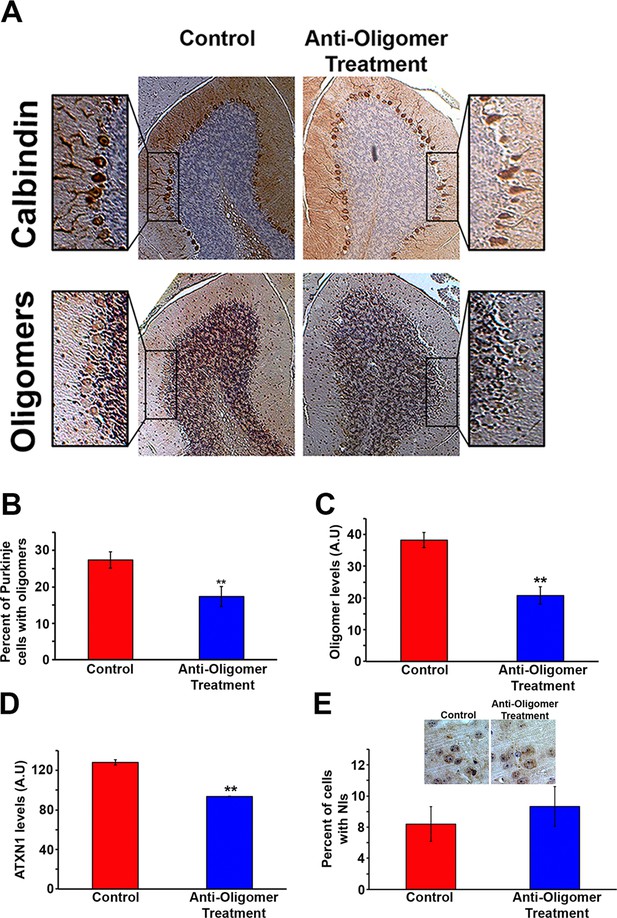
Anti-oligomer immunotherapy decreases pathology in vivo.
(A) Histological staining of PCs (Calbindin, top panels) and Oligomers (A11, bottom panel) of control (IgM) and treated (F11G3) Atxn1154Q/+ mice. Adjacent sections were used for comparison. (B) Quantification of (A), showing the percentage of PCs with oligomers. Data are represented as mean ± SEM., and ** denotes p<0.01, Student’s T-test. (C) ELISA for oligomer levels (A11) in the cerebellum of treated mice and the control group. Data are represented as mean ± SEM., and ** denotes p<0.01, Student’s T-test. (D) ELISA for ATXN1 (11750) levels in the cerebellum of treated mice and the control group. Data are represented as mean ± SEM., and ** denotes p<0.01, Student’s T-test. (E) Immunotherapy in the cortex of Atxn1154Q/+ mice produced no significant change in the number of cells with nuclear inclusions (NIs, stained with 11750 antibody).
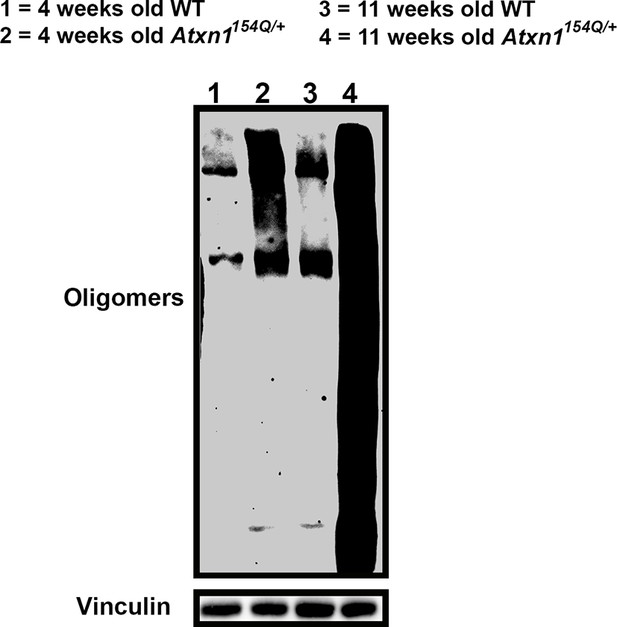
Oligomers are detected in Atxn1154Q/+ cerebellum at four weeks of age.
Western blot against F11G3 to detect oligomers in cerebella from four (lanes 1 and 2) and 11 week (panels 3 and 4) old WT and Atxn1154Q/+ mice. Oligomers are detected in Atxn1154Q/+ at four weeks, but the amount is modest in comparison with eleven weeks of age Atxn1154Q/+.
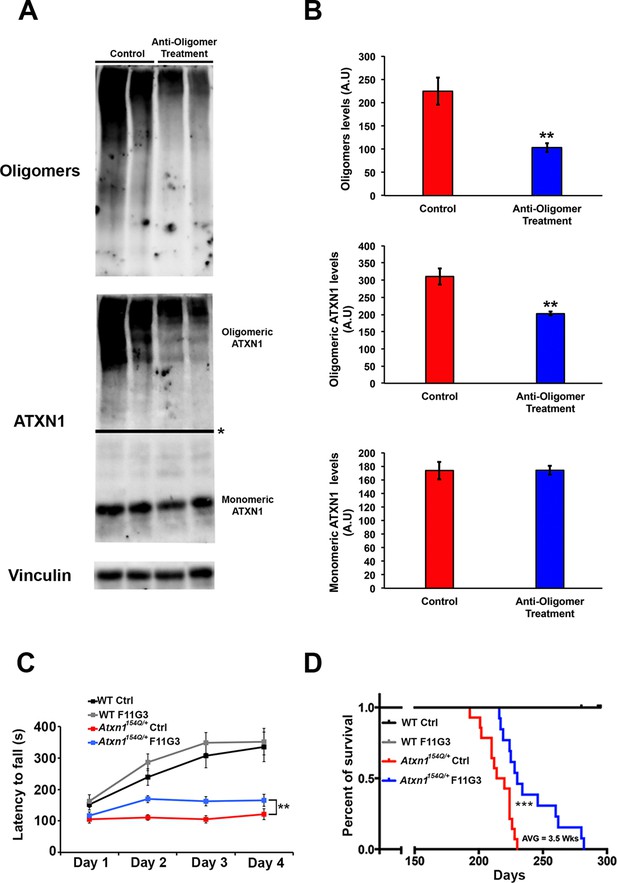
Anti-oligomer immunotherapy improves motor deficits and survival in vivo.
(A–B) Western blot detecting ATXN1 oligomers (F11G3, top panel) and ATXN1 monomer (11750, middle panel) in the cerebellum of Atxn1154Q/+following immunotherapy (anti-oligomer or control). * in (A) indicated change in exposure of the membrane. (B) Quantification of relative levels of oligomeric and monomeric ATXN1 from (A). ** denotes p<0.01, Student’s T-test. (C) Rotarod assay in all treatment groups over a four-day period (four trials per day, averaged) 6 weeks following onset of immunotherapy (mice 10 weeks of age). n = 12 per genotype; ** denotes p<0.01, ANOVA followed by Tukey’s post hoc test. (D) Kaplan-Meier survival curve shows that animals treated with anti-oligomer immunotherapy (blue line) lived, on average, 3.5 weeks longer than control animals (red line). No death was observed in WT mice receiving immunotherapy (black and grey lines). *** denotes p<0.001, Log-rank (Mantel-Cox) test. n = 12 per genotype.





