Identity of neocortical layer 4 neurons is specified through correct positioning into the cortex
Figures
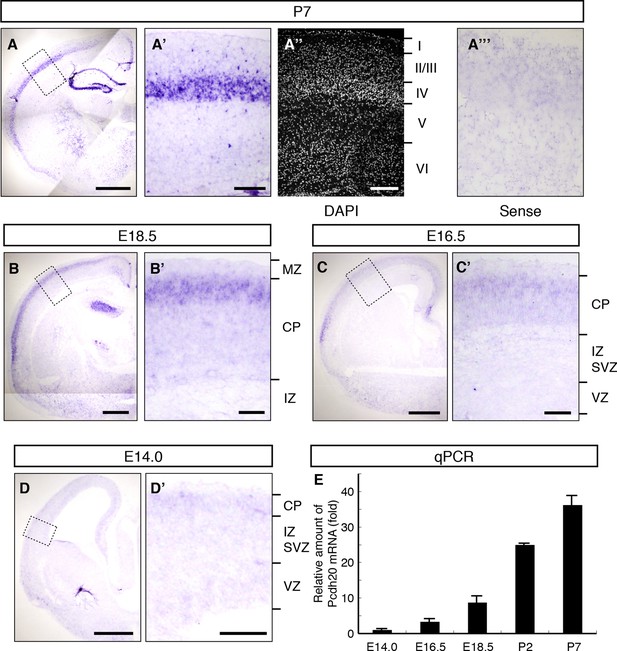
Expression of Pcdh20 mRNA in the developing neocortex.
(A–D) In situ hybridization for Pcdh20 was performed in the E14.0, E16.5, E18.5 and P7 neocortex. The boxed regions in A–D are shown at higher magnification in A’–D’. Nuclear staining with DAPI of the section adjacent to A’ shows the laminar structure of the neocortex (A”). No layer-specific signals were detected with the sense probe in the P7 neocortex (A”’). Expression of Pcdh20 was weak in the E14.0 and E16.5 neocortex, but was clearly evident in the E18.5 neocortex; strong expression was observed in the P7 brain. (E) Quantitative RT-PCR analysis was performed at the indicated stages using Pcdh20-specific primers. Values are means ± SEM of three biological replicates. Scale bars, 1 mm (A); 500 µm (B–D); 200 µm (A’, A”); 100 µm (B’–D’). CP, cortical plate; IZ, intermediate zone; MZ, marginal zone; SVZ, subventricular zone; VZ, ventricular zone.
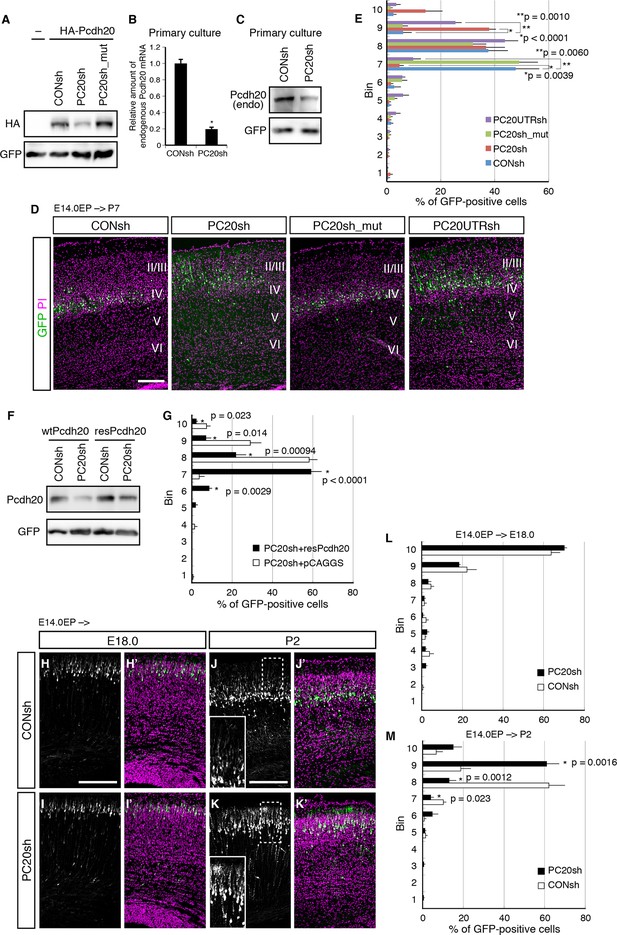
Pcdh20 is required for correct positioning of “future L4 neurons”.
(A) 293T cells were transfected with a control vector (CONsh), an shRNA vector targeting Pcdh20 mRNA (PC20sh), or PC20sh_mut (which harbours point mutations in PC20sh) together with an HA-tagged Pcdh20 expression vector and a GFP expression vector. The cells were subjected to immunoblotting with antibodies to HA and GFP. (B) CONsh or PC20sh vector together with GFP vector was introduced on E14.0 cortices by in utero electroporation. Two days later, the cortices were removed, dissociated and cultured for 4 days in vitro. The GFP-positive cells were FACS sorted, and the amounts of Pcdh20 mRNA were then analyzed by RT-qPCR. The Pcdh20 levels were normalized by the expression of β-actin. Values are means ± SEM of three biological replicates. (C) CONsh or PC20sh vector together with GFP vector was introduced in dissociated E15.5 cortical cells by electroporation. Four days later, the cells were subjected to immunoblotting with antibodies to Pcdh20 and GFP. (D, E) CONsh, PC20sh, PC20sh_m, or PC20UTRsh vector together with a GFP vector was electroporated into the lateral ventricle of E14.0 embryos, then, the P7 brains were fixed and analyzed. The sections were counterstained with propidium iodide (PI, magenta). Most GFP-positive cells in the control experiment were located in L4, while the cells carrying the PC20sh or PC20UTRsh vector were located mainly in L2/3. Results of quantitative analyses of D are presented in E (n = 6 CONsh, n = 6 PC20sh, n = 5 PC20sh_mut, n = 5 PC20UTRsh). Details are described in Materials and Methods. (F) 293T cells were transfected with CONsh or PC20sh together with wild-type Pcdh20 (wtPcdh20) or a resistant form of Pcdh20 harboring mutations in the PC20sh-targeting site (resPcdh20). The cells were subjected to immunoblotting with antibodies to Pcdh20 and GFP. (G) PC20sh vector together with resPcdh20 was injected, and the brains were analyzed as in E. Results of quantitative analyses of Figure 2—figure supplement 1A are presented in G (n = 4 PC20sh+pCAGGS, n = 5 PC20sh+resPcdh20). (H–K) CONsh (H, J) or PC20sh (I, K) vector together with a GFP vector was electroporated into E14.0 brains, then, E18.0 (H, I) and P2 (J, K) brains were analyzed. The sections were counterstained with PI (magenta). The boxed regions are shown at higher magnification in the insets (J, K). Note that most GFP-positive cells with the PC20sh vector migrated normally, but were malpositioned in the P2 brains. (L, M) Quantitative data from E18.0 (H, I) (n = 4 CONsh, n = 4 PC20sh) and P2 (J, K) (n = 4 CONsh, n = 4 PC20sh) brains are presented. Scale bars, 200 µm (D, H, J).
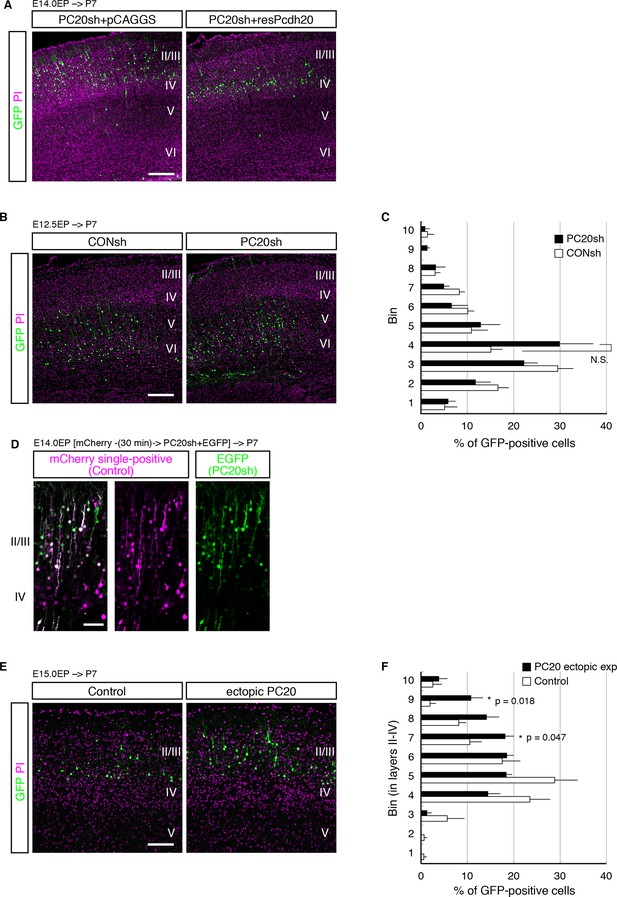
Pcdh20 is required for correct positioning of “future L4 neurons”.
(A, B) PC20sh vector together with resPcdh20 was injected, and the brains were analyzed as in Figure 2D. (B, C) Experiments were done as in Figure 2D and E, except that electroporation was performed into E12.5 brains. (D) Sequential electroporation of GFP, followed 30 min later by that of PC20sh and mCherry, was performed. Control cells are magenta in color in the merged figure (indicating that they were positive for only mCherry), while the PC20sh-transfected cells are green. (E, F) Experiments were done as in Figure 2D and E, except that electroporation was performed into E15.0 brains. Scale bars, 200 µm (A, B, E); 100 µm (D).
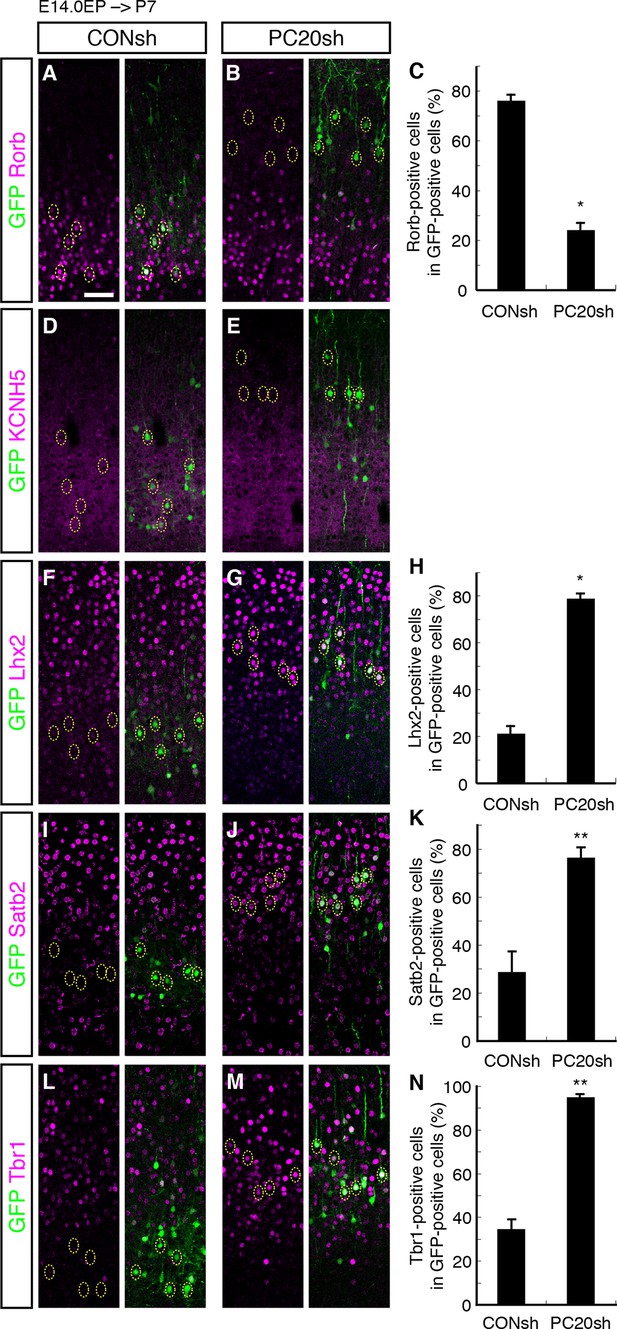
Acquisition of L2/3 molecular features of “future L4 neurons” induced by Pcdh20 knockdown.
(A–N) CONsh or PC20sh vector together with a GFP vector was electroporated into E14.0 brains, then, P7 brains were fixed and analyzed. The sections were immunostained for Rorb (A, B), KCNH5 (D, E), Lhx2 (F, G), Satb2 (I, J), and Tbr1 (L, M). The images at low magnification are shown in Figure 3—figure supplement 1. The results of quantitative analyses for Rorb (C), Lhx2 (H), Satb2 (K), and Tbr1 (N) are presented (n = 4 CONsh, n = 4 PC20sh). Pcdh20-knockdown cells showed diminished expressions of Rorb and KCNH5, but acquired expressions of Lhx2, Satb2 and Tbr1. *p < 0.0001, **p < 0.005. Scale bar, 50 µm (A).
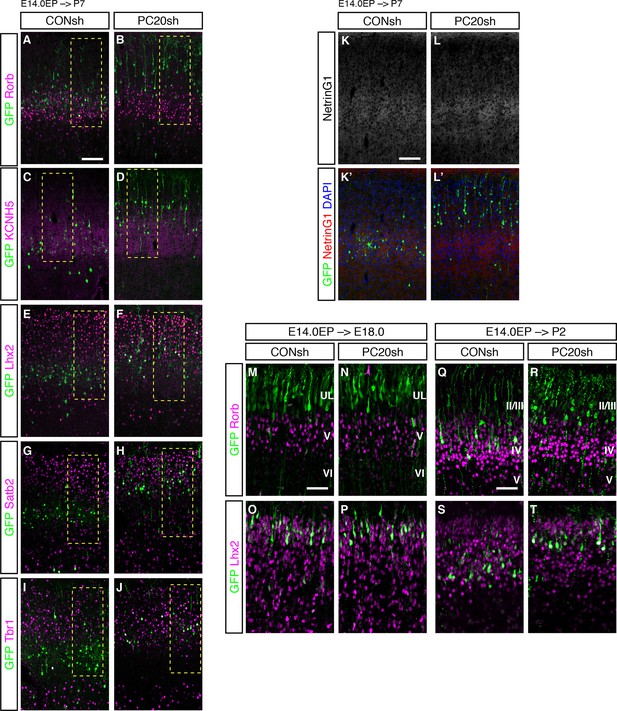
Pcdh20 knockdown resulted in “future L4 neurons” acquiring L2/3 characteristics.
(A–J) Images shown in Figure 3A–M are presented at low magnification. (K, L) CONsh or PC20sh vector together with a GFP vector was electroporated into E14.0 brains, then, P7 brains were fixed and analyzed. The sections were counterstained with DAPI (blue) and immunostained for NetrinG1 (red). (M–T) CONsh or PC20sh vector together with a GFP vector was electroporated into E14.0 brains, then, E18.0 (M–P) and P2 (Q–T) brains were analyzed. The sections were immunostained for Rorb and Lhx2. Scale bars, 100 µm (A, K); 50 µm (M, Q).
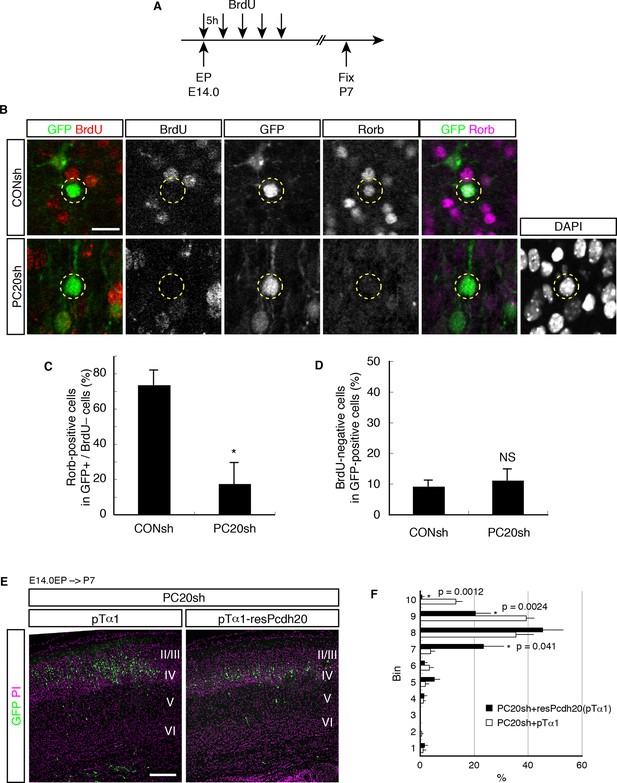
Postmitotic function of Pcdh20 regulates the development of L4 neurons.
(A) The experimental procedure to identify postmitotic cells from VZ cells. (B) The brains treated as in A were immunostained for BrdU, GFP and Rorb. Typical images of this experiment are shown. (C) The percentages of Rorb-positive cells among the GFP-positive and BrdU-negative cells were analyzed. The Pcdh20-knockdown cells showed loss of expression of Rorb even in a postmitotic population. (D) The percentages of BrdU-negative cells among GFP-positive cells are presented. The percentages of the BrdU-negative cells were not affected by the knockdown of Pcdh20. (E, F) PC20sh vector together with the empty vector (pTα1) or Tα1 promoter-driven resPcdh20 expression vector (pTα1-resPcdh20) was electroporated into E14.0 brains, and P7 brains were analyzed. Results of quantitative analyses of E are presented in F. *p = 0.0056; NS, no significance (p = 0.62). Scale bars, 20 µm (B); 200 µm (E).
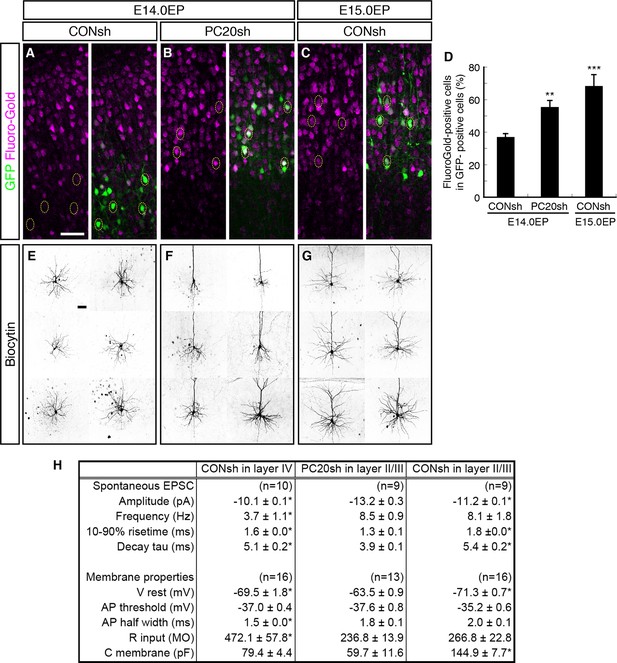
Alterations of axon projections and electrophysiological properties by Pcdh20 knockdown.
(A–D) CONsh (A, C) or PC20sh (B) vector together with a GFP vector was electroporated into E14.0 (A, B) or E15.0 (C) brains. Fluoro-Gold was injected at P7 into the cortex contralateral to the side of electroporation, then, the P10 brains were fixed and analyzed. The images at low magnification are shown in Figure 4—figure supplement 1. Results of quantitative analyses of A–C are presented in D (n = 4 each group). (E–G) CONsh (E, G) or PC20sh (F) vector together with a GFP vector was electroporated into E14.0 (E, F) or E15.0 (G) brains. The cell morphologies were then analyzed in P14–19 brains by injection of biocytin and sequential visualization with avidin-Cy3. The remaining cells are presented in Figure 4—figure supplement 2A–C. Pcdh20-knockdown neurons possessed thick apical dendrites, a characteristic morphologic feature of L2/3 neurons, but not of L4 neurons. (H) Effects on electrophysiological properties by Pcdh20 knockdown. Experiments were performed as described in E–G, and the electrophysiological properties were analyzed at P14-19. *p < 0.05 (compared with the Pcdh20-knockdown brains electroporated on E14.0), **p < 0.005, ***p = 0.0204 (compared with the control brains electroporated on E14.0). Scale bars, 50 µm (A, E).
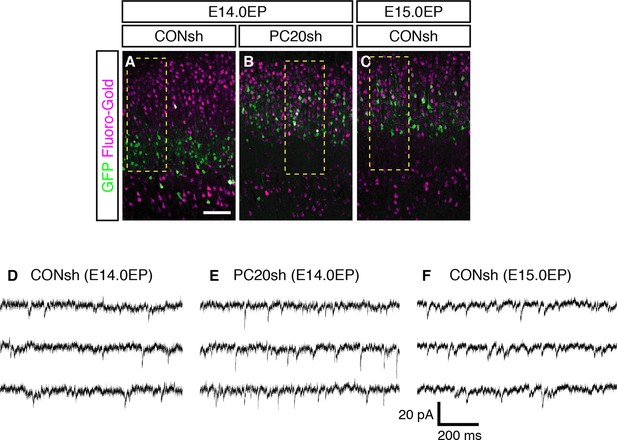
sEPSCs in Pcdh20-knockdown cells and control L2/3 and L4 cells.
(A–C) Images shown in Figure 4A–C are presented at low magnification. (D–F) Examples of sEPSCs described in Figure 4H are presented. Scale bar, 100 µm (A).
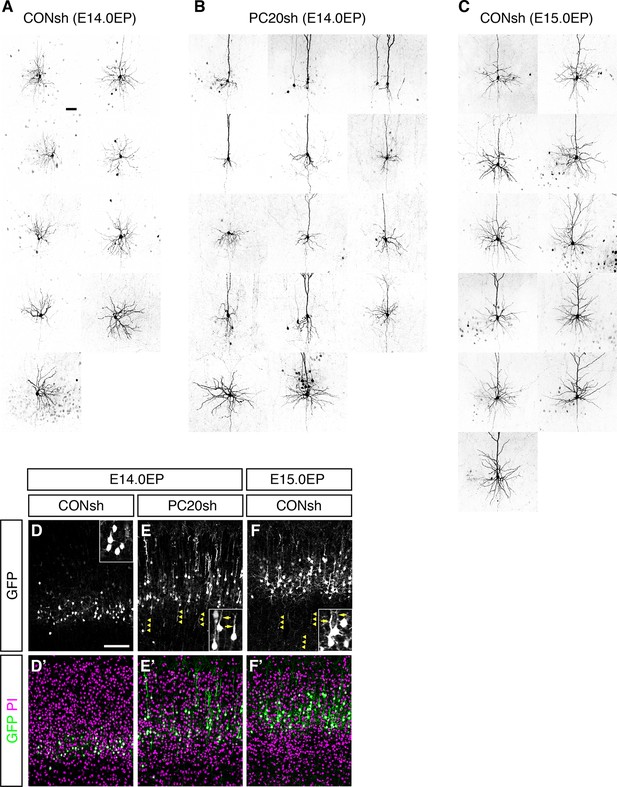
Morphological changes of “future L4 neurons” induced by knockdown of Pcdh20.
(A–C) The remaining cells in Figure 4E–G are presented. (D–F) CONsh (D, F) or PC20sh (E) vector together with a GFP vector was electroporated into E14.0 (D, E) or E15.0 (F) brains, then, P7 brains were fixed and analyzed. The sections were counterstained with PI (magenta). High-magnification images are presented in the insets. The Pcdh20-knockdown cells possessed apical dendrites and axons, which are characteristics of pyramidal neurons in L2/3 of the neocortex. Scale bars, 50 µm (A); 100 µm (D).
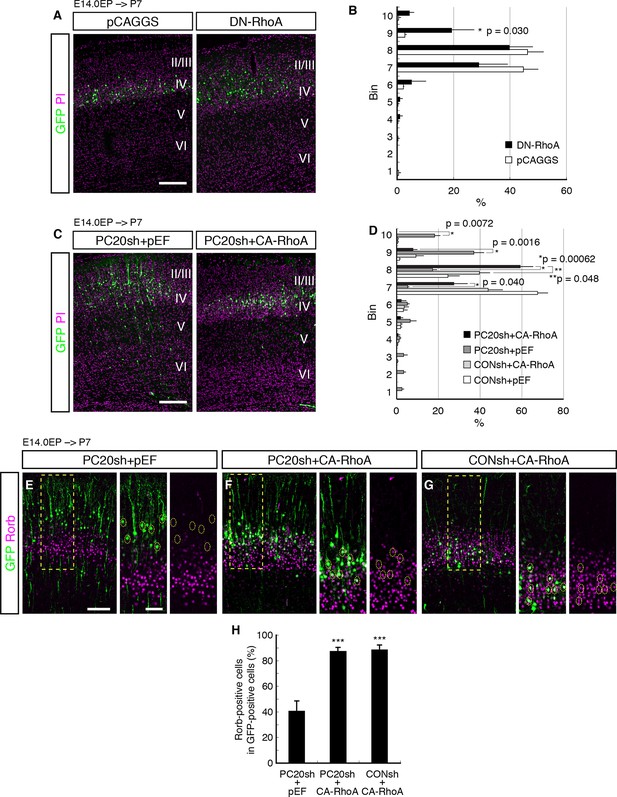
Involvement of RhoA in the Pcdh20-dependent development of L4 neurons.
(A, B) The empty vector (pCAGGS) or dominant-negative RhoA expression vector (DN-RhoA) was electroporated into E14.0 brains, and P7 brains were analyzed. Results of quantitative analyses of A are presented in B (n = 4 pCAGGS, n = 4 DN-RhoA). Many of the DN-RhoA-expressing cells were located in L2/3. (C, D) The indicated vectors were electroporated into E14.0 brains, and P7 brains were analyzed. Results of quantitative analyses of c are presented in D (n = 4 each group). Constitutively active RhoA (CA-RhoA) rescued the malpositioning caused by the knockdown of Pcdh20. (E–H) The sections prepared from the brains in C were immunostained for Rorb and GFP. Results of quantitative analyses of E–G are presented in H (n = 4 PC20sh+pEF, n = 4 PC20sh+CA-RhoA, n = 5 CONsh+CA-RhoA). Active RhoA restored the specification failure induced by Pcdh20 knockdown. ***p < 0.005. Scale bars, 200 µm (A, C); 100 µm (E on the left); 50 µm (E on the right).
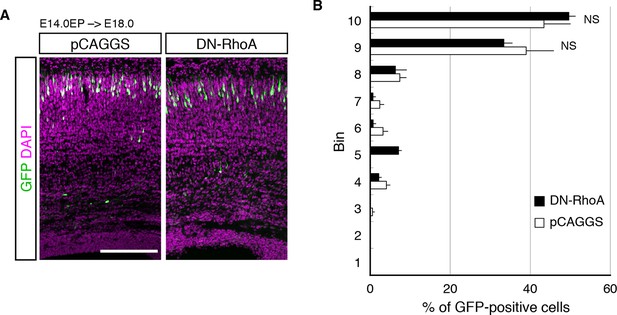
DN-RhoA did not change the cell positioning in early stages.
(A, B) The empty vector (pCAGGS) or dominant-negative RhoA expression vector (DN-RhoA) was electroporated into E14.0 brains, and E18.0 brains were analyzed. Results of quantitative analyses of A are presented in B (n = 4 pCAGGS, n = 4 DN-RhoA). Scale bars, 200 µm (A).
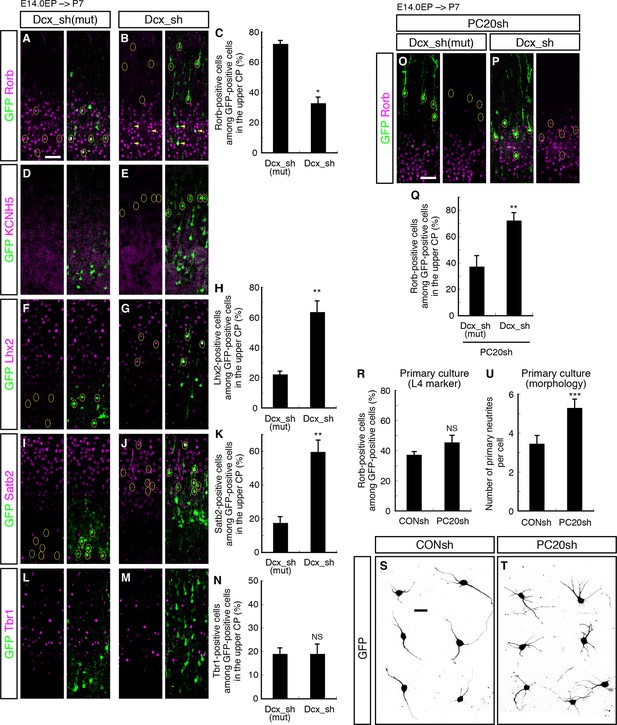
Failure of subtype specification of the cells positioned ectopically in L2/3 by knockdown of Dcx.
(A–N) Control (Dcx_sh(mut), harboring point mutations in Dcx_sh) or Dcx_sh vector together with a GFP vector was electroporated into E14.0 brains, then, P7 brains were fixed and analyzed. The sections were immunostained for Rorb (A, B), KCNH5 (D, E), Lhx2 (F, G), Satb2 (I, J) and Tbr1 (L, M). The images at low magnification are shown in Figure 6—figure supplement 1. The results of quantitative analyses for Rorb (C), Lhx2 (H), Satb2 (K) and Tbr1 (N) are presented (n = 4 Dcx_sh(mut), n = 4 Dcx_sh). Dcx-knockdown cells exhibited some of the phenotypes observed in the Pcdh20 knockdown experiment. (O–Q) Dcx_sh(mut) or Dcx_sh vector together with the PC20sh vector was electroporated into E14.0 brains, then, P7 brains were fixed and analyzed. The sections were immunostained for Rorb and GFP (O, P). The results of quantitative analyses are presented in Q (n = 4 Dcx_sh(mut)+PC20sh, n = 4 Dcx_sh+PC20sh). The cells showing recovery of positioning in L4 following introduction of the PC20sh and Dcx_sh vectors also showed recovery of Rorb expression. (R–U) CONsh or PC20sh vector together with a GFP vector was electroporated into E14.0 brains, then, E18.0 brains were dissociated. The neurons were cultured for 4 days (R) or 2 days (S–U) and immunostained for GFP and Rorb (R) or GFP (S–U). Results of quantitative analyses are presented in R (n =3 CONsh, n =3 PC20sh) and U (n = 22 CONsh, n = 21 PC20sh). *p < 0.0001, **p < 0.02, ***p < 0.005. Scale bars, 50 µm (A, O); 20 µm (S).
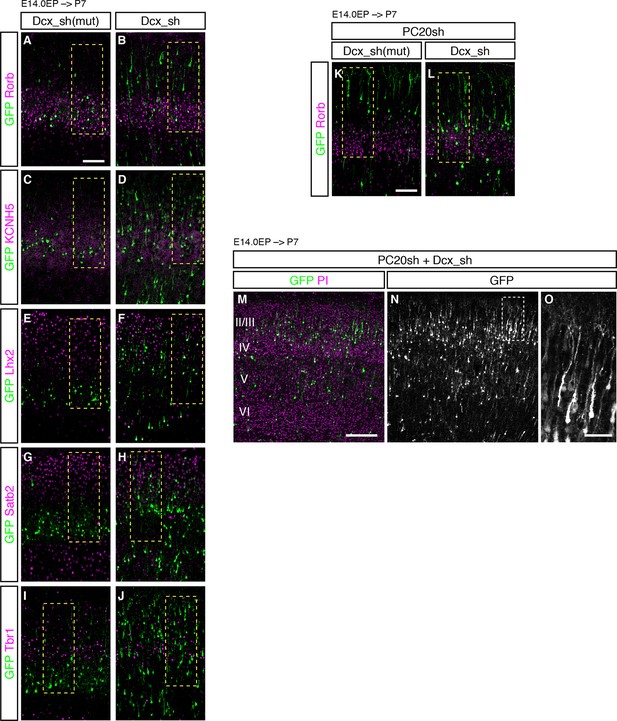
Failure of subtype specification of the cells positioned ectopically in L2/3 by knockdown of Dcx.
(A–J) Images shown in Figure 6A–M are presented at low magnification. (K, L) Images shown in Figure 6O and P are presented at low magnification. (M–O) Expression of PC20sh and Dcx_sh resulted in both phenotypes. Dcx_sh vector together with PC20sh and a GFP vector was electroporated into E14.0 brains, then, P7 brains were fixed and analyzed. The sections were counterstained with PI (magenta). The boxed region in N is shown at a higher magnification in O. The neurons were positioned in a random manner throughout the neocortex. In addition, some of them showed well-developed apical dendrites, a feature of Pcdh20-knockdown cells, suggesting that both RNAi vectors were effective. Scale bars, 100 µm (A, K); 200 µm (M); 50 µm (O).
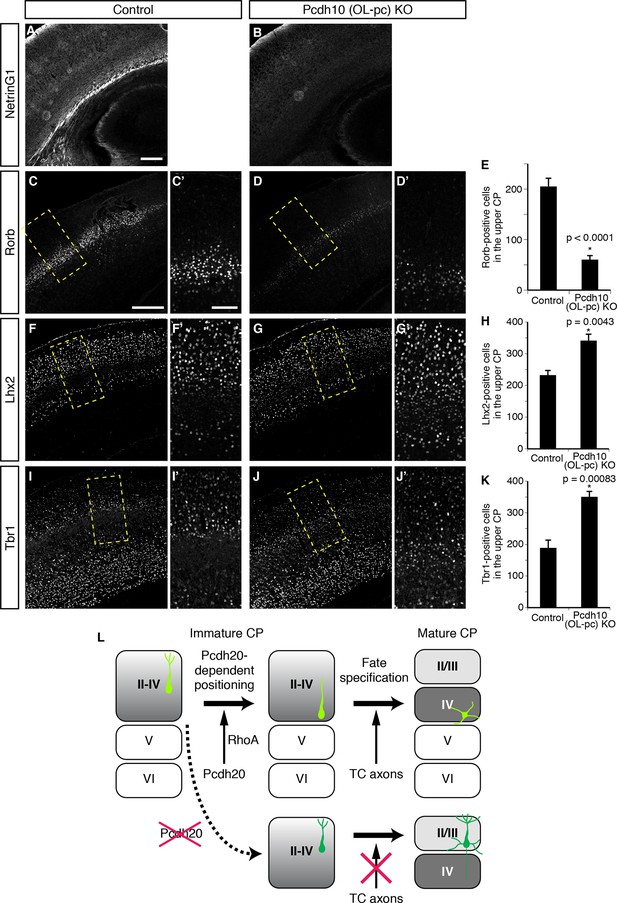
Requirement of TCAs for correct differentiation of L4 neurons.
(A–K) The sections prepared from Pcdh10 (OL-pc) knockout mice were immunostained for NetrinG1 (A, B), Rorb (C, D), Lhx2 (F, G) and Tbr1 (I, J). The boxed regions are shown at higher magnification in the columns on the right. Typical images of the P7 cortices were shown. Quantitative analyses were performed on the P3 cortices (E, H, K) (n = 6 Control, n = 5 Pcdh10 (OL-pc) KO for Rorb; n = 4 Control, n = 4 Pcdh10 (OL-pc) KO for Lhx2; n = 4 Control, n = 5 Pcdh10 (OL-pc) KO for Tbr1). (L) A model for L4 specification suggested by our study is presented. We propose that the identities of the superficial layer neurons are not completely specified before the neurons eventually come to reside beneath the marginal zone (left). After radial migration, the future L4 neurons become positioned in the lower part of the superficial cortical plate in a Pcdh20-dependent manner (upper middle). This positioning enables the future L4 neurons to come in contact with TCAs, which appears to be required for further specification and maturation of the L4 neurons (upper right). In the absence of Pcdh20, the neurons reside in the upper part and are unable to receive signals from TCAs (lower row). Scale bars, 200 µm (A, C); 100 µm (C’).





