TANGO1 recruits ERGIC membranes to the endoplasmic reticulum for procollagen export
Figures
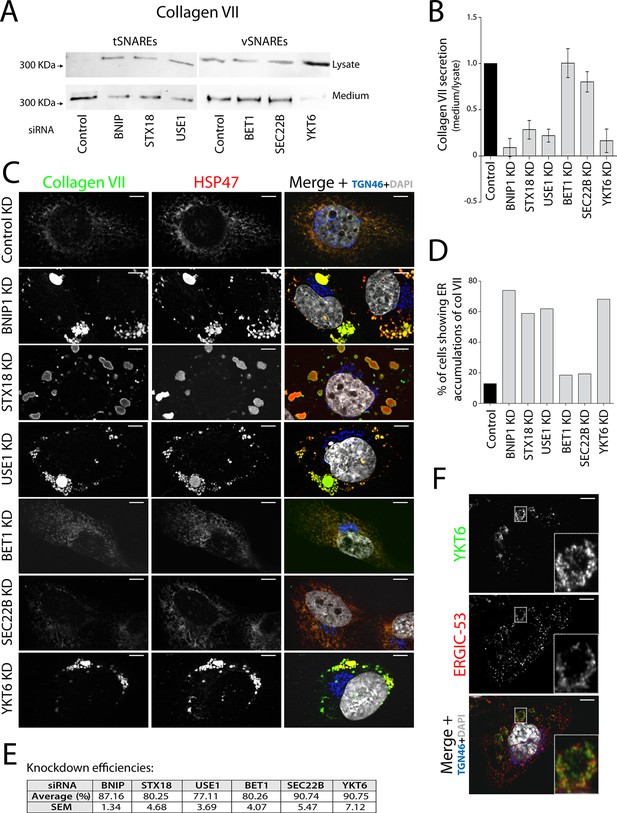
V- and t-SNAREs required for procollagen VII export from the ER RDEB/FB/C7 cells were transfected with siRNAs directed against BNIP1, STX18, USE1, BET1, SEC22B, YKT6 or a scrambled siRNA.
(A) Collagen VII secretion was measured by western blotting and probing for collagen VII in RDEB/FB/C7 cell lysates and supernatants collected for 20 hr in the presence of ascorbic acid. In three independent experiments, intensities of the collagen VII signal in the lysate and the supernatant was recorded by densitometry. (B) The ratio of external vs. internal Collagen VII was normalized to quantify secretion in control cells as 1; Error bars: standard error of the mean (SEM). Differences in secretion between control and each knockdown were statistically significant, as determined by the Mann Whitney U test. (C) siRNA-treated RDEB/FB/C7 cells were seeded on coverslips and 20 hr after addition of ascorbate, cells were fixed and visualized with the indicated antibodies and DAPI by fluorescence microscopy (scale bars: 10 µm). (D) The percentage of cells that accumulate intracellular Collagen VII was determined by counting at least 20 cells in five random fields. (E) Efficiency of knockdown of each SNARE by quantifying mRNA or protein levels. (F) YKT6 is localised to vesicular structures, many of which colocalise with ERGIC-53. Manders’ overlap coefficient 0.705 ± 0.118.
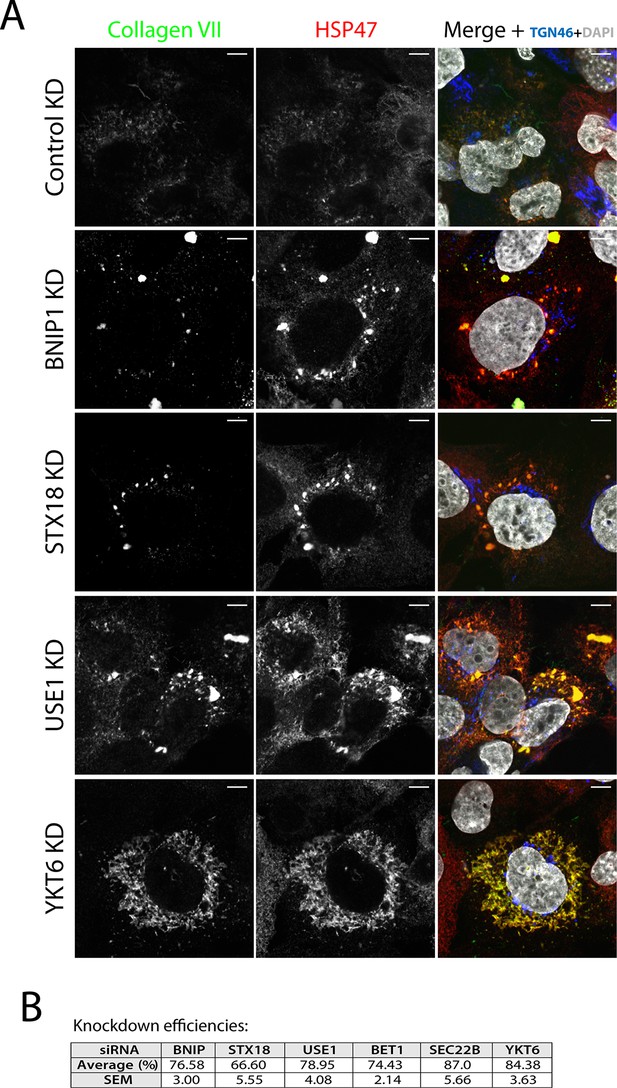
SNAREs Syntaxin 18, BNIP1, USE1 and YKT6 are required for procollagen VII export from the ER in Het-1A cells.
(A) Het-1A cells transfected with siRNA as described in the methods section were incubated with ascorbate for 20h, fixed with methanol and immunostained with for collagen VII, HSP47 and TGN46. Accumulation of procollagen VII in the ER was observed after depletion of these SNAREs. (B) Efficiency of knockdown of each SNARE by quantifying mRNA or protein levels.
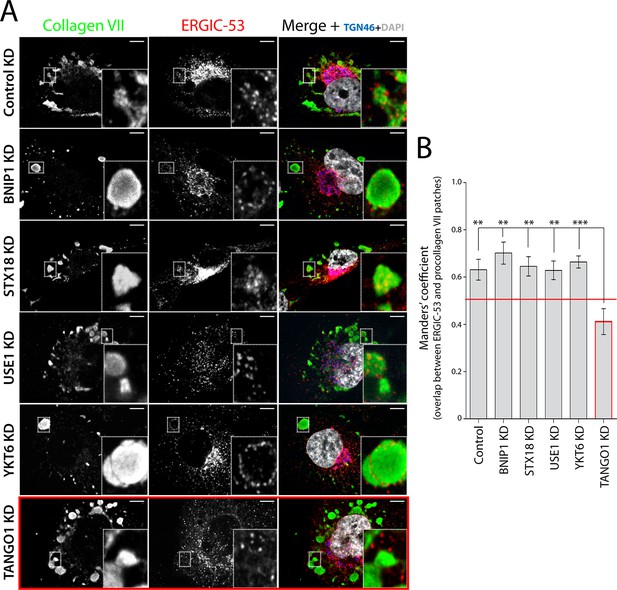
TANGO1-dependent recruitment of ERGIC membranes to ER-retained patches of procollagen VII.
(A) Procollagen VII was accumulated in the ER of RDEB/FB/C7 cells that were either incubated at 15°C to accumulate secretory cargo in the ER and then released at 37°C for five minutes (control) or transfected with siRNA oligos to specifically deplete ER t-SNAREs BNIP1, Syntaxin 18 or USE1, the v-SNARE YKT6, or TANGO1. The cells were immunostained to monitor the localisation of ERGIC membranes and procollagen VII. Zooms are of 4x and scale bars correspond to 10 µm. (B) Colocalisation quantifications were calculated by Manders’ correlation coefficient by measuring the overlap between the green and the red channels. Error bars reflect the standard error of the mean (SEM) of more than 20 cells from at least three independent experiments. **p<0.01, ***p<0.001.
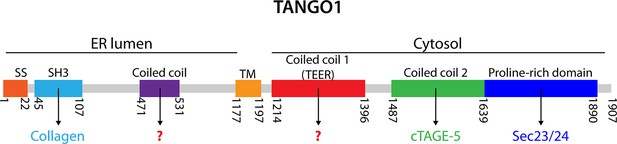
TANGO1 domains.
A schematic representation of human TANGO1 domain organization and its known interactors. SS, signal sequence; SH3, Src homology 3-like domain that is required for binding procollagen VII (Saito et al., 2009); TM, transmembrane domain; Coiled coil 2 that is known to interact with cTAGE5 (Saito et al., 2011), and the proline-rich domain (PRD) that interacts with Sec23/24 (Saito et al., 2009).
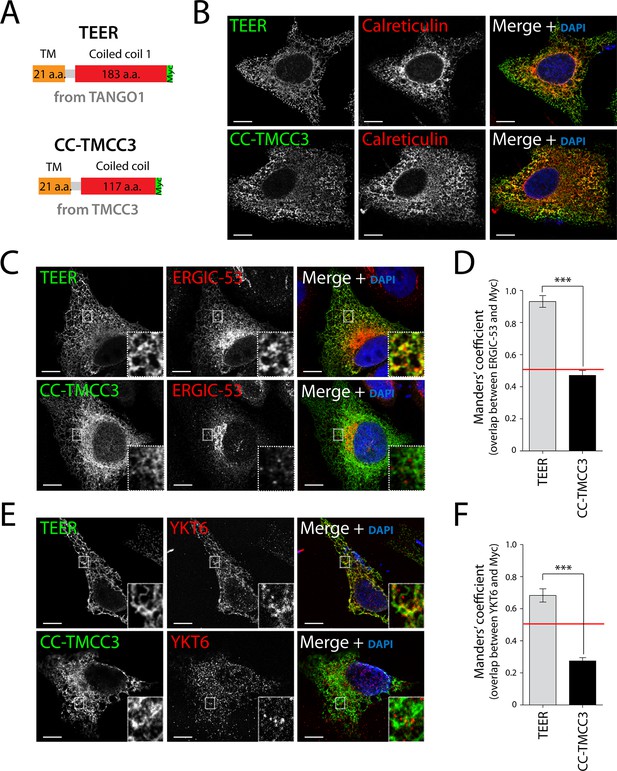
Expression of TEER recruits YKT6-containing ERGIC membranes to the ER.
(A) A scheme depicting the coding regions of the two constructs - TEER-Myc (residues 1177–1396 of TANGO1, tagged with Myc) and CC-TMCC3-Myc (residues 282–437 of TMCC3, tagged with Myc). HeLa cells were seeded on coverslips and fixed 48 hr after transfection with the constructs. For visualization, upon fixation and permeabilisation, cells were stained with DAPI (blue) and immunostained for Myc (green). The red channel shows immunostaining for (B) the ER marker calreticulin; for (C) the transmembrane protein ERGIC-53; and (E) the v-SNARE YKT6 (cells were washed after permeabilisation to remove the cytosolic proteins prior to fixation as described in materials and methods). In the case of CC-TMCC3, YKT6 signal intensity was increased 3 times during image acquisition. Zooms are of 5x and scale bars correspond to 10 µm. (D-F) Colocalisation quantifications were calculated by Manders’ correlation coefficient by measuring the overlap between the green and the red channels. Error bars reflect the standard error of the mean (SEM) of more than 20 cells from at least three independent experiments. ***p<0.001.
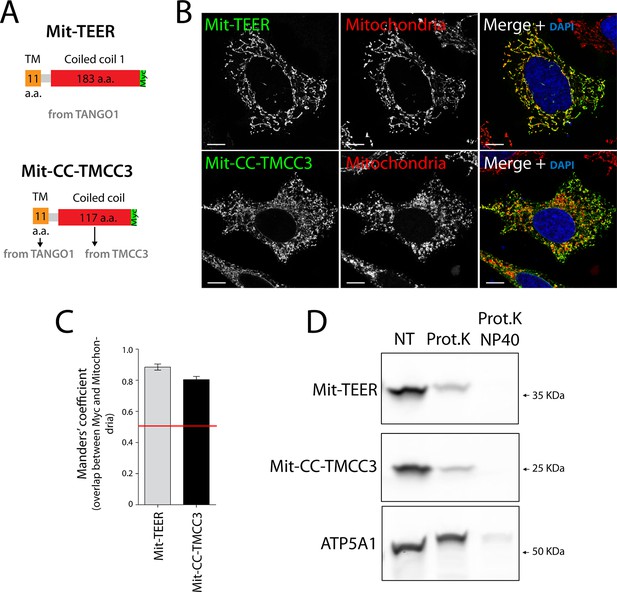
The sequence and topology of mitochondrially targeted TEER (Mit-TEER) and CC-TMCC3 (Mit-CC-TMCC3).
(A) A scheme depicting the coding regions of the two constructs for mitochondrial targeting - Mit-TEER-Myc (residues 1187–1396 of TANGO1, including 11 amino acids of its transmembrane domain, tagged with Myc) or Mit-CC-TMCC3-Myc (the same 11 amino acids of TANGO1’s transmembrane domains and residues 282–398 of TMCC3, tagged with Myc). (B) HeLa cells were seeded on coverslips and transfected with Mit-TEER-Myc or Mit-CC-TMCC3-Myc for 48 hr. To visualize mitochondria (red), cells were either incubated with the Mito-tracker dye for 30 min in serum-free medium before fixation or stained with an anti-ATP5A1 antibody after fixation and permeabilisation. They were also stained with DAPI (blue) and immunostained for Myc (green). (C) Colocalisation quantifications were calculated by Manders’ correlation coefficient by measuring the overlap between the green and the red channels. Error bars reflect the standard error of the mean (SEM) of more than 20 cells from at least three independent experiments. Scale bars correspond to 10 µm. ***P <0.001. (D) Mitochondrial membranes were isolated from HeLa cells transfected with either Mit-TEER or Mit-CC-TMCC3 and split into three fractions. The first was kept as a control (NT); proteinase K (0.1mg/ml) (Prot.K) was added to the second fraction; and to the third fraction, both proteinase K and the detergent NP40 (at a final concentration of 1%) were added (Prot.K/NP40). Samples were incubated on ice for one hour and western blotted with an anti-Myc antibody, to detect Mit-TEER and Mit-CC-TMCC3, and an anti-ATP5A1, a subunit of the ATP synthase, localized to the mitochondrial inner membrane. The loss of Mit-TEER/Mit-TMCC3 but not of ATP5A1, in the absence of detergent, indicated that these constructs are exposed to the cytoplasm.
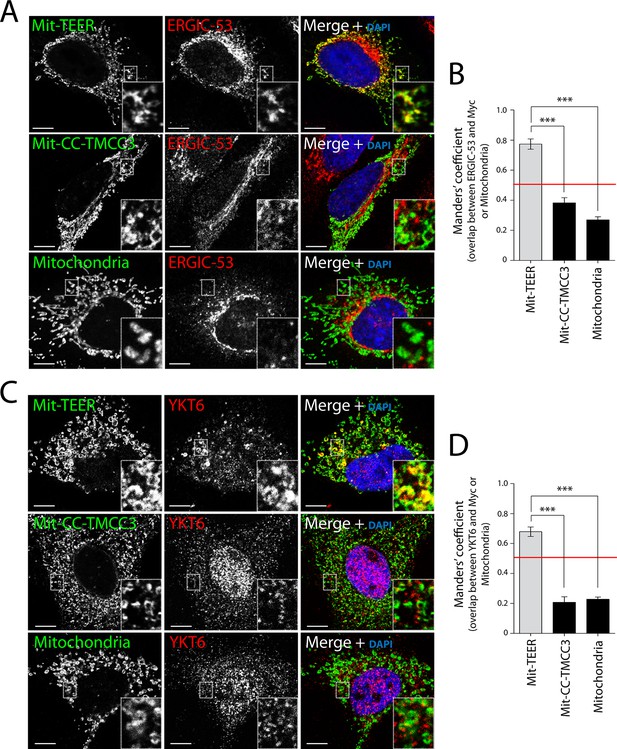
Recruitment of YKT6-containing ERGIC membranes to TEER expressed at the mitochondria (Mit-TEER).
HeLa cells were seeded on coverslips and transfected with Mit-TEER-Myc or Mit-CC-TMCC3-Myc for 48 hr. For visualisation, cells were stained with DAPI (blue) and immunostained for Myc (green), and were also immunostained for (A) the transmembrane protein ERGIC-53 (red); and (C) the v-SNARE YKT6 (red) (in cells washed after permeabilisation to remove cytosolic protein followed by fixation and immunostaining). In the case of CC-TMCC3 and untransfected control, YKT6 signal intensity was increased 3 times during image acquisition. Zooms are of 5x and scale bars correspond to 10 µm. (B-D) Quantification of colocalisation was calculated by Manders’ correlation coefficient by measuring the overlap between the green and the red channels. Error bars reflect the standard error of the mean (SEM) of more than 20 cells counted from at least three independent experiments. ***P <0.001.
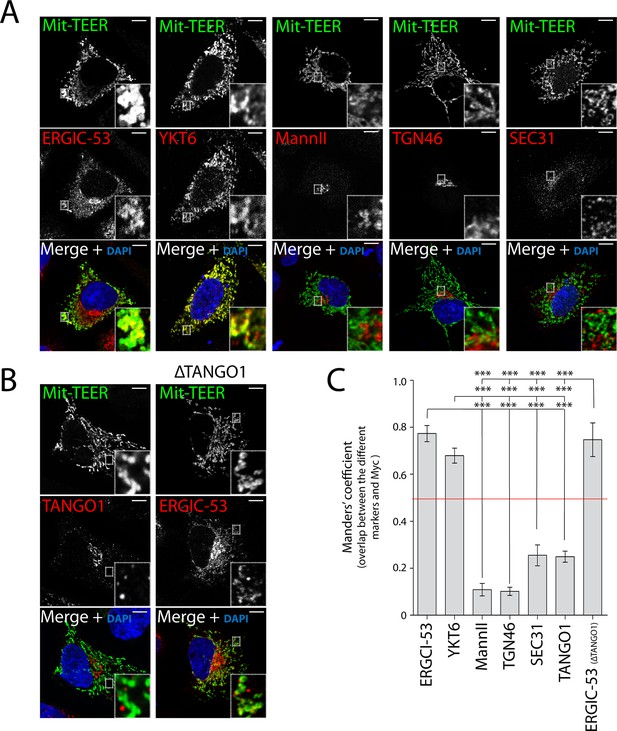
TEER recruits ERGIC membranes, but not Golgi membranes or COPII components HeLa cells were seeded on coverslips and 48 hr after transfection with Mit-TEER-Myc they were fixed and permeabilised.
(A) For visualisation, cells were stained with DAPI (blue) and immunostained for Myc (green). The red channel shows immunostaining for ERGIC-53; YKT6 (after cytosol washout); early and late Golgi markers (Mannosidase II and TGN46); and the COPII coat component Sec31. (B) Left panel - HeLa cells transfected with Mit-TEER-Myc as before were visualised following DAPI staining (blue), immunostaining for Myc (green) and TANGO1 (red). Right panel - HeLa cells depleted of TANGO1 protein by CRISPR procedure were transfected with Mit-TEER-Myc as before and visualized following DAPI staining (blue) and immunostaining for Myc (green) and ERGIC-53 (red). Zooms are of 5x and scale bars correspond to 10 µm. (C) Quantifications of colocalisation was calculated by Manders’ correlation coefficient by measuring the overlap between the green and the red channels. Error bars reflect the standard error of the mean (SEM) of more than 10 cells counted from at least three independent experiments. ***P <0.001.
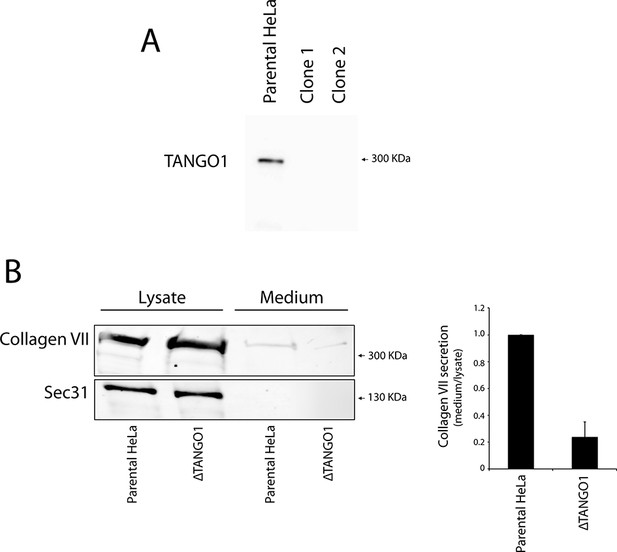
Generation of TANGO1-deficient HeLa by CRISPR/Cas9.
(A) Immunoblot of the parental HeLa cell line and two Tango1 CRISPR knock-out HeLa clones. Protein lysates were separated on a 6% SDS-PAGE and transferred onto a nitrocellulose membrane and probed by western blotting with an anti-TANGO1 antibody. (B) HeLa cells lacking TANGO1 (ΔTANGO1) were transfected with a FLAG-epitope-tagged collagen VII plasmid for 28 hr and then incubated for 20 hr with fresh medium containing ascorbate to promote procollagen VII export from the ER. The cell lysate and medium from ΔTANGO1 HeLa cells and parental HeLa cells were western blotted for collagen VII. In three independent experiments, intensities of the collagen VII-FLAG signal in the lysate and the supernatant was recorded by densitometry and the ratio of external vs. internal collagen VII-FLAG was normalized to quantify secretion in control cells as 1; Error bars: standard error of the mean (SEM).
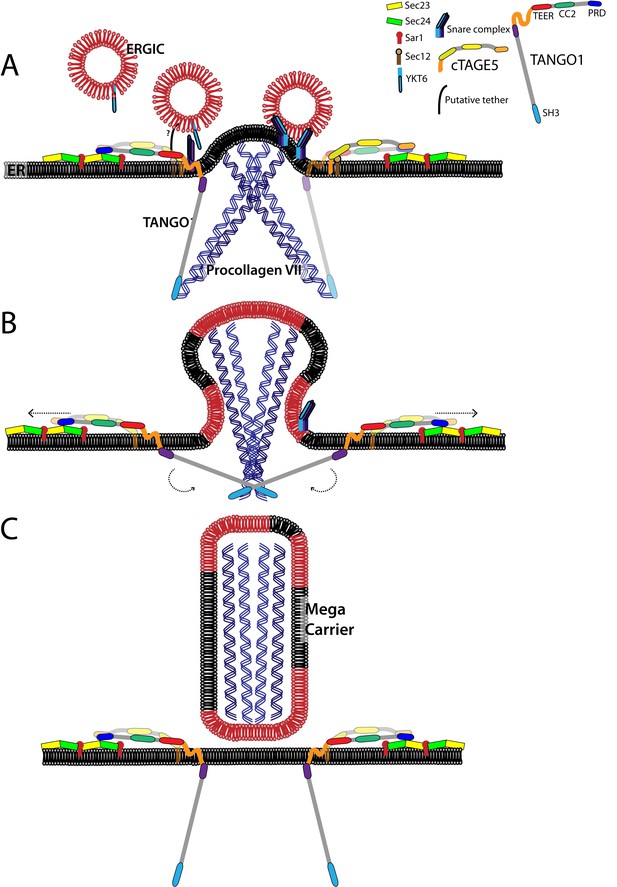
A model for the generation of procollagen VII containing mega-carriers.
(A) A pool of ERGIC membranes fuse to the ER exit site enriched in procollagen VII. This fusion is mediated by the t-SNARES BNIP1, Syntaxin18 and USE1; and the v-SNARE YKT6. The TEER domain of TANGO1 acts as a tether binding to an unidentified protein to recruit ERGIC membranes. (B) Binding of the proline-rich domain of TANGO1 on the cytoplasmic side to Sec23/24, prevents Sec13/31 recruitment, allowing continuous fusion of ERGIC membranes that result in the growth of the nascent bud into a mega-bud that is enriched in procollagen VII. (C) The SH3-like domain of TANGO1 dissociates from procollagen VII, which triggers the separation of the proline-rich domain from Sec23/24. This activates fission, to release the procollagen VII filled mega-carrier from the ER. TANGO1 remains at the ER exit site and not exported with the outgoing procollagen VII.





