Precise assembly of complex beta sheet topologies from de novo designed building blocks
Figures
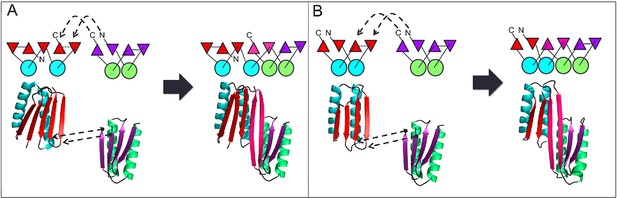
Generation of protein domains with single extended beta sheets by inserting one beta sheet containing protein into another.
(A) Insertion of a ferrrodoxin domain (purple) into TOP7 (red). (B) Insertion of one ferrodoxin domain into another. In both cases, two beta strands from each partner (red and purple) are concatenated to form the central strand pair of the fusion protein (pink).
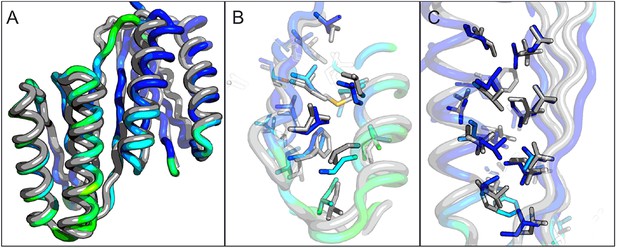
Comparison of the crystal structure of ferredoxin-TOP7 fusion to design model.
(A) Backbone superposition of the crystal structure of ferredoxin-TOP7 (4KYZ, chain A) with the design model. The backbones of the two proteins are nearly identical. (B, C) The core sidechain packing in the ferrodoxin-TOP7 fusion is very similar in the crystal structure and design model both in the insert (B) and host (C) domains. The crystal structure is colored by B-factor and the design model is in gray.
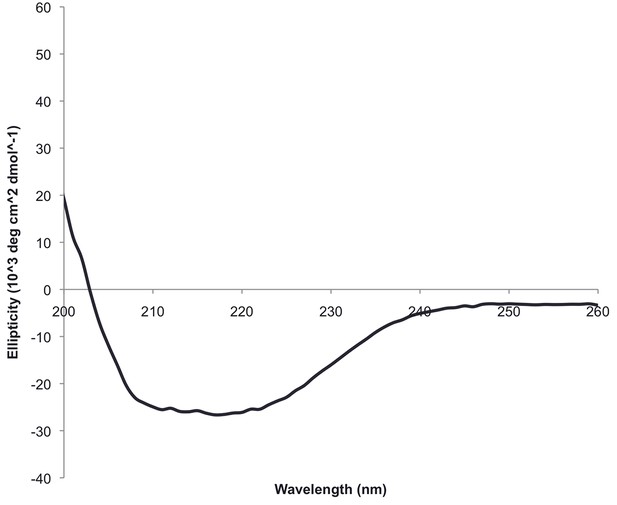
The circular dichroism spectrum of ferrodoxin-TOP7 has the shape expected for an alpha/beta protein.
https://doi.org/10.7554/eLife.11012.005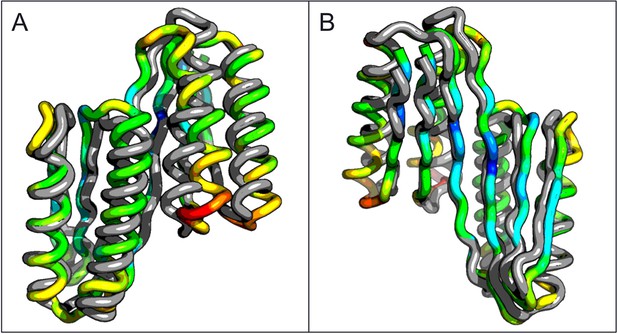
Comparison of the crystal structure of the ferredoxin-ferredoxin fusion to the design model.
The crystal structure (5CW9) aligns well with the design model over both the helices (A) and the fused beta sheet (B).
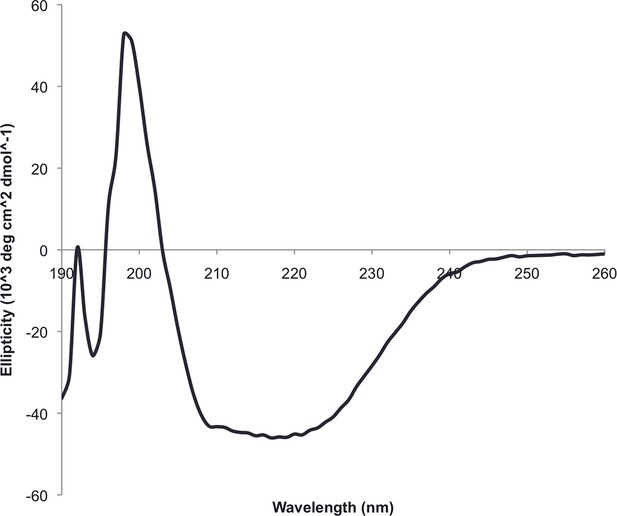
Circular dichroism spectra of ferrrodoxin-ferrodoxin at 25°C.
https://doi.org/10.7554/eLife.11012.007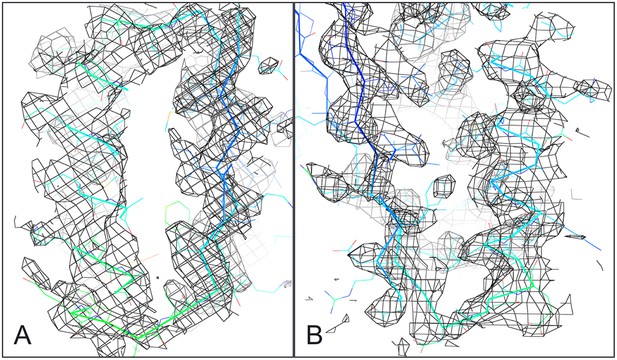
Ferredoxin-Ferredoxin 2Fo-Fc omit map superimposed with crystal structure shows core packing of host (A) and insert (B) domains.
https://doi.org/10.7554/eLife.11012.008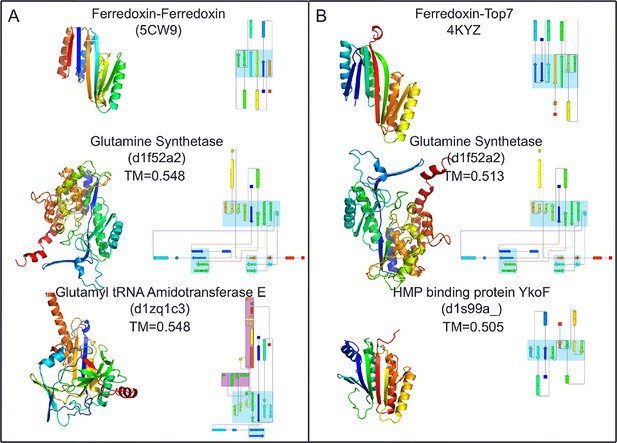
Top two SCOP domain structural homologues for Fd-Top7 (A) and Fd-Fd (B) designed domain found in TM-align searches.
Ribbon diagrams are shown on left, the strand connectivity, at the right. The beta strand connectivity is quite different in the designs than in these closest structural matches.
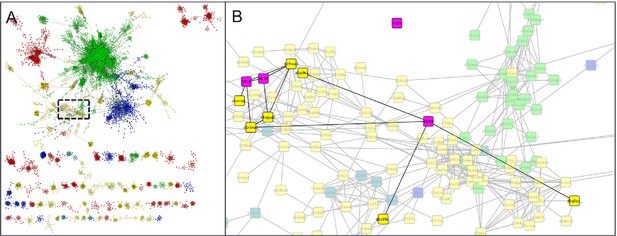
Parent domain PDB structures (2KL8, 1QYS) and daughter designed folds (5CW9,4KYZ) (pink) mapped into the α+β region of the SCOP domains network of Nepomnyachi et al. (A) and zoomed region (B) highlighting parent, designed, and first neighbor folds.
https://doi.org/10.7554/eLife.11012.010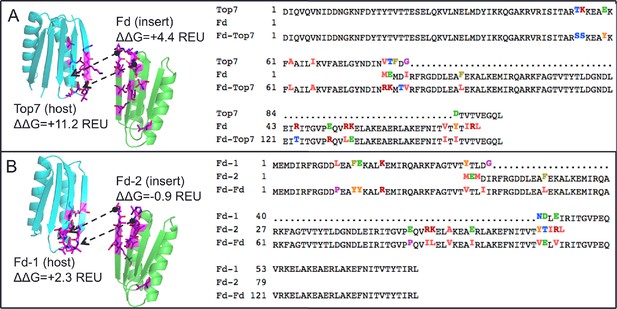
Neutral drift mutant models, relative changes to predicted free energy of folding in REU (Rosetta Energy Units), and multiple sequence alignment of parent and designed sequences, showing mutations in ferredoxin-top7 (A) and ferredoxin-ferredoxin (B).
https://doi.org/10.7554/eLife.11012.011Additional files
-
Supplementary file 1
Crystallographic data.
- https://doi.org/10.7554/eLife.11012.012





