Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway
Figures
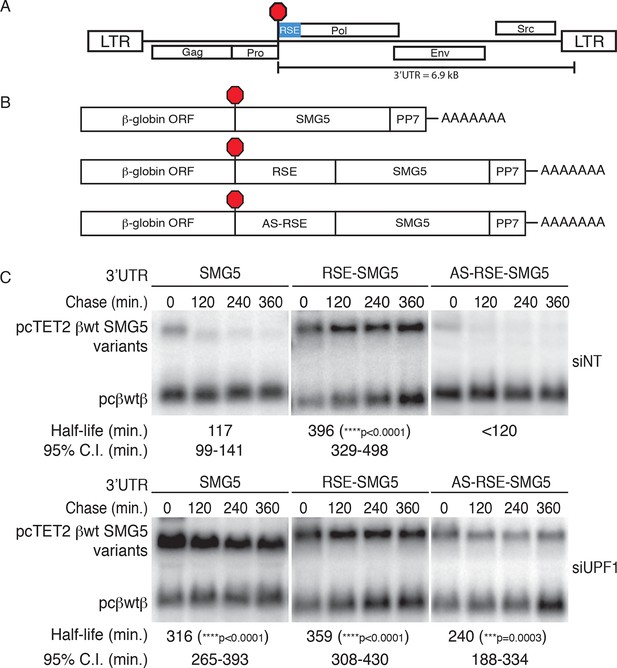
The RSE protects reporter mRNA from NMD in mammalian cells.
(A) Schematic of the Rous sarcoma proviral genome. The RSE is located immediately downstream of the gag stop codon. (B) Schematic of tet-regulated β-globin reporter mRNA constructs used in RNA decay assays. The RSE sequence (middle) and a control sequence, the antisense RSE (AS-RSE) sequence (bottom), were inserted into reporter mRNAs containing the β-globin gene and the human SMG5 3’UTR (top). (C) Decay assays of reporter mRNAs containing the wild-type SMG5 3’UTR or variants supplemented with RSE or AS-RSE sequences. 293 Tet-off cells were treated with non-targeting siRNA (siNT; upper panel) or UPF1 siRNA (siUPF1; lower panel). Constructs encoding the indicated tet-regulated transcripts were co-transfected with the constitutively expressed wild-type β-globin reporter (pcβwtβ; bottom bands). Remaining RNA levels at indicated time points were normalized to levels of the wild-type β-globin transfection control. Half-lives and 95% confidence intervals were obtained from 3 independent experiments (***p<0.001; ****p<0.0001 in two-tailed ANCOVA analysis when compared to pcTET2-βwt-SMG5). Rapid decay of AS-RSE mRNAs to background levels in siNT samples precluded accurate quantification of decay rate. See also Figure 1—figure supplements 1 and 2.
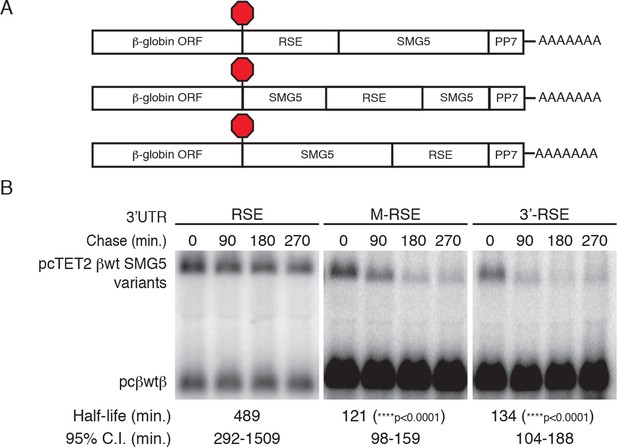
RSE protective activity is position-dependent.
(A) Schematic of tet-regulated β-globin reporter mRNA constructs used in RNA decay assays. RSE sequence was inserted in reporter mRNAs either upstream, in the middle, or downstream of the SMG5 3’UTR. (B) Decay assays of reporter mRNAs containing the RSE at different positions in the 3'UTR. Constructs encoding the tet-regulated transcripts described in A (pcTET2-βwt-RSE-SMG5, pcTET2-βwt-MRSE-SMG5, or pcTET2-βwt-3’RSE-SMG5; top bands) were co-transfected with the constitutively expressed wild-type β-globin reporter (pcβwtβ; bottom bands) in HeLa Tet-off cells for decay analysis. Remaining RNA levels at indicated time points were normalized to levels of the wild-type β-globin transfection control. Half-lives and 95% confidence intervals were obtained from 3 independent experiments (p-values from two-tailed ANCOVA analysis when compared to pcTET2-βwt-RSE-SMG5).
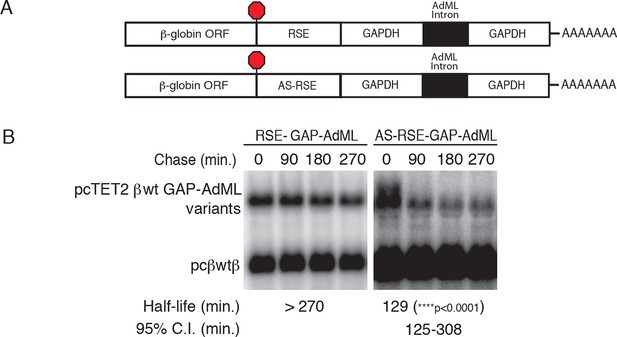
The RSE protects reporter mRNAs from EJC-stimulated NMD
(A) Schematic of tet-regulated β-globin reporter mRNA constructs used in RNA decay assays. RSE or antisense RSE (AS-RSE) sequences were inserted in reporter mRNAs upstream of a previously-characterized version of the GAPDH-derived 3’UTR engineered to contain the adenovirus major-late intron (AdML intron; Singh et al., 2008), leading to EJC-stimulated NMD. (B) Decay of AdML intron-containing mRNAs in HeLa Tet-off cells. Half-lives and 95% confidence intervals were derived from linear regression of semi-log plots of normalized RNA abundances from three independent experiments (p-values from two-tailed ANCOVA analysis comparing the AS-RSE mRNAs to the RSE mRNAs).
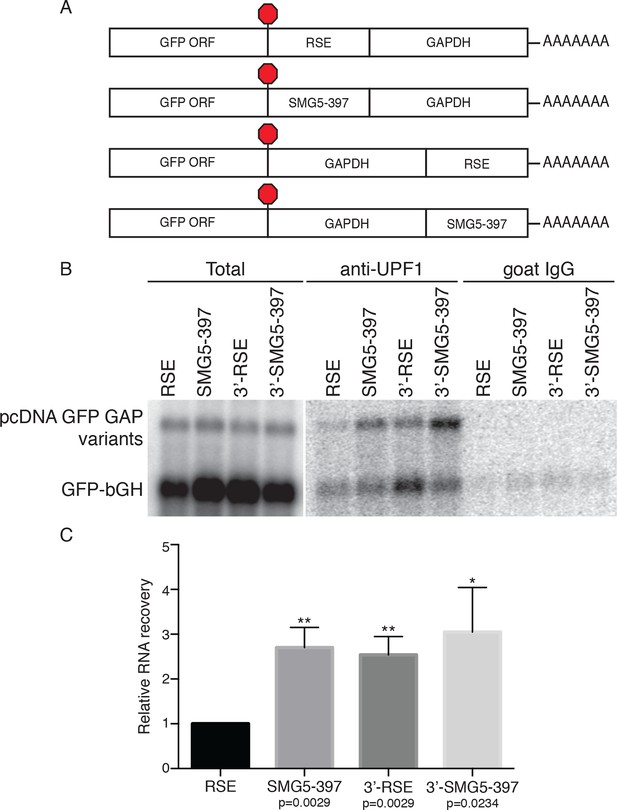
The RSE reduces UPF1 association with 3'UTRs in a position-dependent manner.
(A) Schematic of reporter mRNA constructs used in immunoprecipitation assays. RNAs containing the GFP ORF and an artificial NMD-inducing 3’UTR comprising a portion of the human GAPDH ORF and the GAPDH 3’UTR (Singh et al., 2008) were modified to contain the RSE or the SMG5-397 sequence 5’ or 3’ of the GAPDH sequence. (B) Upf1 is reduced on 3'UTRs containing a TC-proximal RSE. Plasmids expressing the indicated mRNAs were co-transfected in 293 cells with a construct expressing the GFP ORF followed by the bovine growth hormone (bGH) polyadenylation signal. Endogenous UPF1 was immunoprecipitated from transfected cells, and co-purifying mRNAs were analyzed by northern blot. Bulk goat IgG was used as a non-specific interaction control. (C) Quantification of relative RNA recovery upon UPF1 immunoprecipitation, normalized to the recovery of co-transfected GFP-bGH mRNAs. The amount of RSE-containing RNA recovered was set to 1. Error bars indicate ± SD; n = 3 (*p<0.05; **p<0.01 in two-tailed Student’s t-tests when compared to RSE recovery). See also Figure 2—figure supplements 1 and 2.
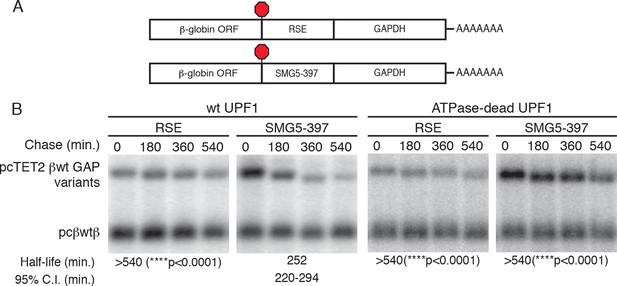
The RSE protects reporter mRNAs containing a GAPDH-derived NMD-sensitive 3’UTR.
(A) Schematic of tet-regulated β-globin reporter mRNA constructs used in RNA decay assays. RSE or SMG5-397 sequences were inserted in reporter mRNAs upstream of the artificial GAPDH-derived 3’UTR (Singh et al., 2008). (B) Decay assays of reporter mRNAs containing the RSE or the first 397 nt of the SMG5 3’UTR (SMG5-397) sequence in cells expressing either WT UPF1 (left) or an ATPase-dead UPF1 (right). Constructs encoding the tet-regulated transcripts described in (A) were co-transfected with the constitutively expressed wild-type β-globin reporter (pcβWTβ; bottom bands) in HeLa Tet-off cells. Half-lives and 95% confidence intervals were derived from linear regression of semi-log plots of normalized RNA abundances from three independent experiments (p-values from two-tailed ANCOVA analysis comparing the indicated mRNAs to the SMG5-397 control in the presence of wild-type UPF1).
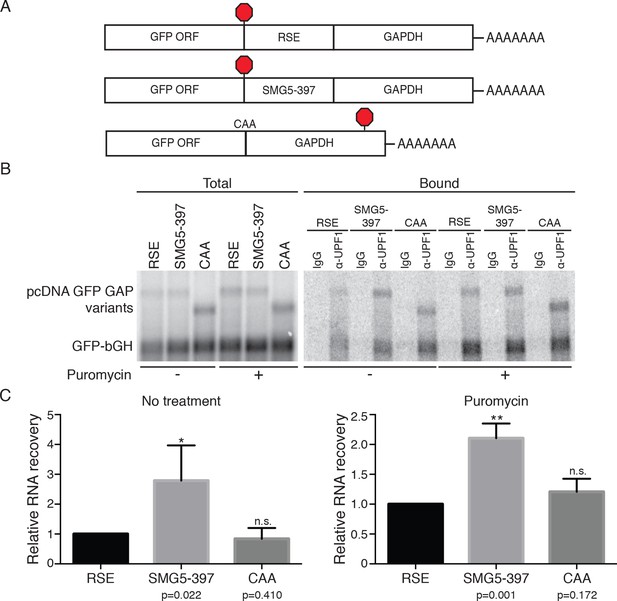
RSE inhibition of UPF1 binding is independent of translation.
(A) Schematic of GFP reporter mRNA constructs used in immunoprecipitation assays. RNAs containing the GFP ORF and an artificial NMD-inducing 3’UTR comprising a portion of the human GAPDH ORF and the GAPDH 3’UTR (Singh et al., 2008) were modified to contain the RSE (top) or the SMG5-397 sequence (middle). As a control to illustrate the behavior of mRNAs with short 3’UTRs (untreated condition) or shorter overall length (puromycin treatment), we used constructs without added RSE or SMG5-397 sequence in which the GFP termination codon was changed to CAA (bottom), resulting in termination at the downstream GAPDH stop codon. (B) UPF1 association is reduced on 3'UTRs containing the RSE. Plasmids expressing the indicated mRNAs were co-transfected in 293 cells with a construct expressing the GFP ORF followed by the bovine growth hormone (bGH) polyadenylation signal. Endogenous Upf1 was immunoprecipitated from transfected cells grown in the absence or presence of puromycin (100 μg/mL for 3 hr) as indicated, and co-purifying mRNAs were analyzed by northern blot. (C) Quantification of relative RNA recovery upon UPF1 immunoprecipitation, normalized to the recovery of co-transfected GFP-bGH mRNAs. The amount of RSE-containing RNA recovered was set to 1. Error bars indicate ± SD; n ≥ 3 (*p<0.05, **p<0.01 in two-tailed Student’s t-tests when compared to RSE recovery). Under both untreated and puromycin treated conditions, the RSE caused reduced accumulation of UPF1 on mRNAs relative to the SMG5-397 control.
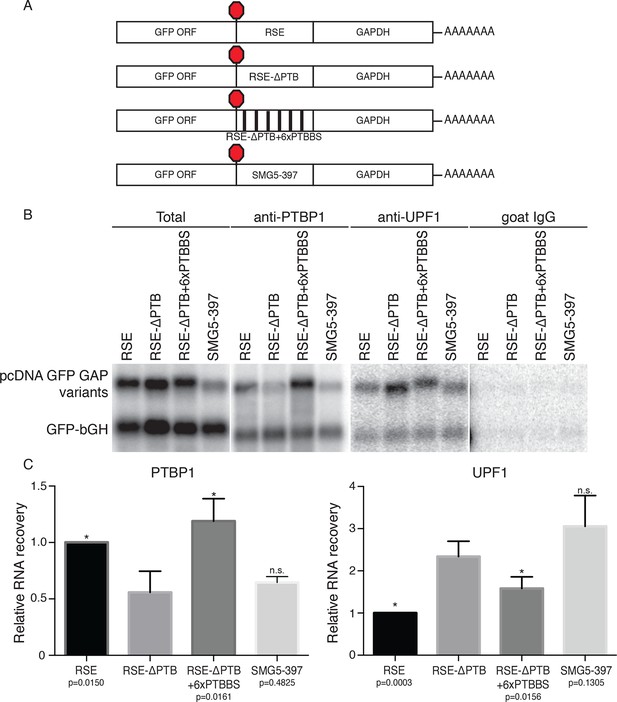
Accumulation of PTBP1 on the 3‘UTR prevents UPF1 binding.
(A) Schematic of reporter mRNA constructs used in immunoprecipitation assays. (B) PTBP1 is reduced on transcripts containing the RSE mutants lacking the putative PTBP1 binding sites. Immunoprecipitations were performed as in Figure 2B. Goat IgG samples were imaged using identical settings to those used for anti-UPF1 samples. (C) Left Panel: Quantification of relative RNA recovery upon PTBP1 immunoprecipitation, normalized to the recovery of co-transfected GFP-bGH mRNAs. Right Panel: Quantification of relative RNA recovery upon UPF1 immunopreciptation, normalized to the recovery of co-transfected GFP-bGH mRNAs. The amount of RSE-containing RNA recovered was set to 1. Error bars indicate ± SD; n ≥ 3 (*p<0.05 in two-tailed Student’s t-tests when compared to RSE-ΔPTB recovery. See also Figure 3—figure supplement 1 and Figure 3—source data 1.
-
Figure 3—source data 1
Table of raw mass spectrometry data.
List of proteins identified in mRNP purifications of PP7-tagged mRNAs containing RSE or antisense (AS) sequence, with numbers of peptides and spectral counts derived from each protein indicated.
- https://doi.org/10.7554/eLife.11155.010
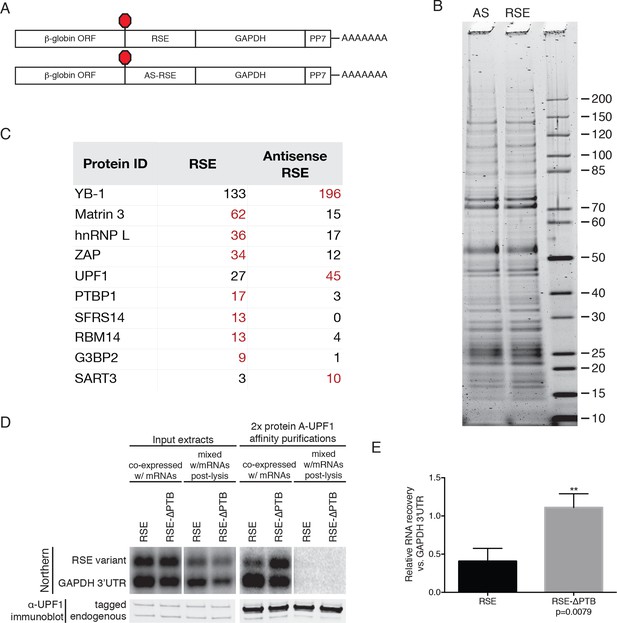
Identification of RSE-interacting proteins by tandem mass spectrometry.
(A) Schematic of β-globin reporter mRNA constructs used for PP7-based affinity purification. The RSE sequence (top) and a control sequence, the antisense RSE sequence (AS-RSE; bottom) were inserted into reporter mRNAs containing the β-globin gene and the GAPDH 3’UTR. (B) mRNP profiles of the transcripts described in (A). Proteins were separated on a 4–12% SDS-PAGE gel and visualized by Krypton Infrared Protein Stain (Pierce). (C) Purified mRNPs were subjected to trypsin digestion and tandem mass spectrometry. Spectral counts from selected proteins enriched in either the RSE-containing sample or the antisense RSE sample are shown. Complete mass spectrometry data are compiled in Figure 3—source data 1. (D) Control for post-lysis reassortment. Extracts from cells in which protein A-tagged UPF1 was co-expressed with the indicated GFP reporter mRNAs or extracts from cells separately expressing protein A-tagged UPF1 and the exogenous mRNAs were used for affinity purification. mRNAs containing the GAPDH artificial 3’UTR were used as recovery controls. Top panels: Northern blots of co-transfected reporter mRNAs. Identical settings were used to image northern blots of input extracts and purified material from co-expressed and mixed samples, respectively. Bottom panels: immunoblotting of mixed extracts with an α-UPF1 antibody indicates equal purification efficiency across all conditions. (E) Relative recoveries of the indicated mRNAs with affinity purified UPF1 were determined, normalized to recovery of mRNAs containing the GAPDH 3’UTR. Error bars indicate ± SD; n = 3 (**p<0.01 in two-tailed Student’s t-test comparing the recovery of RSE-GAPDH to RSE-∆PTB-GAPDH).
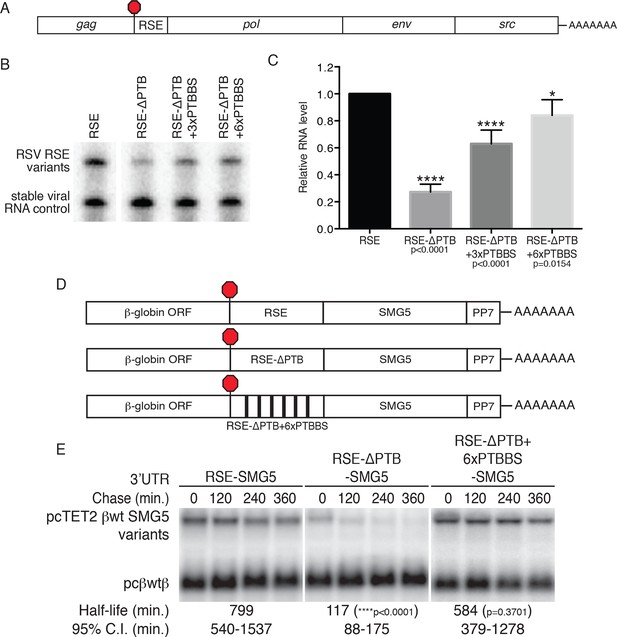
PTBP1 plays an essential role in the RSE’s protective activity against NMD.
(A) Schematic of unspliced RSV RNA expression constructs used for RNA accumulation assays in chicken fibroblasts. (B) RNase protection assays of RSV unspliced viral RNA containing the wt RSE or RSE variants. Experimental and control constructs were co-transfected into CEFs, and total cellular RNAs were harvested 43–48 hr post-transfection. Top band: protected fragment of the probe corresponding to the unspliced viral RNAs from the experimental constructs. Bottom band: stable viral loading control that protects a different sized fragment of the same probe due to a small in-frame deletion. (C) Quantification of RNase protection assays. Levels of the experimental unspliced RSV RNAs (top band) were normalized to levels of the transfection control (bottom band). RNA levels are reported as a fraction of RSV RNA containing wt RSE. Error bars indicate ± SD; n ≥ 5 (*p<0.05; ****p<0.0001 in two-tailed Student’s t-tests when compared to RSE constructs). (D) Schematic of reporter mRNAs containing the β-globin gene, RSE variants (RSE, RSE-ΔPTB, or RSE-ΔPTB+6xPTBBS), and the full-length human SMG5 3‘UTR. (E) Decay assays of the indicated reporter mRNAs. Tet-regulated transcripts (upper bands) were co-transfected with the constitutively expressed wild-type β globin reporter (pcβwtβ; bottom bands) in HeLa Tet-off cells. Levels of tet-regulated reporter mRNAs were normalized to levels of the wild-type β-globin transfection control. Half-lives and 95% confidence intervals were obtained from 3 independent experiments (p-values from two-tailed ANCOVA analyses when compared to pcTET2-βwt-SMG5). See also Figure 4—figure supplement 1.
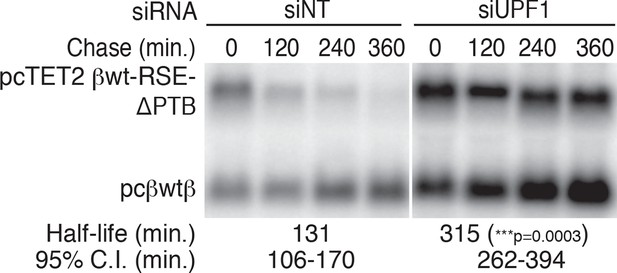
PTPB1 protects reporter mRNAs from NMD.
Decay assays of pcTET2 -βwt-RSE-ΔPTB-SMG5 in HEK293 Tet-off cells treated with non-targeting or anti-UPF1 siRNAs. The reporter constructs (upper band) were co-transfected with the constitutively expressed wild-type β-globin reporter (pcβwtβ; bottom bands) into cells 24 hrs after the siRNAs were introduced to the cells. Levels of tet-regulated reporter mRNAs were normalized to levels of the wild-type β-globin transfection control. Relative remaining RNA levels at indicated time points from three independent experiments were used to calculate half-lives and 95% confidence intervals (***p<0.001 in two-tailed ANCOVA analysis, compared to the half-life of βwt-RSE-ΔPTB-SMG5 in cells treated with non-targeting siRNAs).
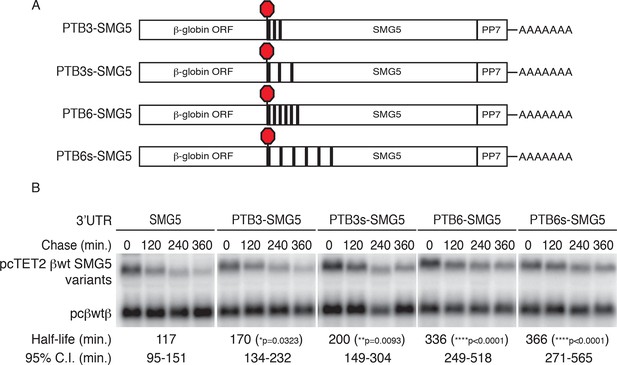
PTBP1 protects NMD-sensitive transcripts from NMD when artificially recruited to the 3‘UTR.
(A) Schematic of tet-regulated β-globin reporter mRNA constructs used in RNA decay assays. Three or six canonical PTBP1 binding sites were inserted into the SMG5 3‘UTR. PTBP1 binding sites were inserted with short linker sequences between each site (PTB3 and PTB6), or spaced at 100 nt intervals within the SMG5 3‘UTR (PTB3s and PTB6s). (B) Decay assays of reporter mRNAs containing PTBP1 binding sites. Constructs encoding the indicated tet-regulated transcripts (top bands) were co-transfected with the constitutively expressed wild-type β-globin reporter (pcβwtβ; bottom bands) in HeLa Tet-off cells. Levels of tet-regulated reporter mRNAs were normalized to levels of the wild-type β-globin transfection control. Half-lives and 95% confidence intervals were obtained from 3 independent experiments (p-values from two-tailed ANCOVA analyses when compared to pcTET2-βwt-SMG5). See also Figure 5—figure supplement 1.
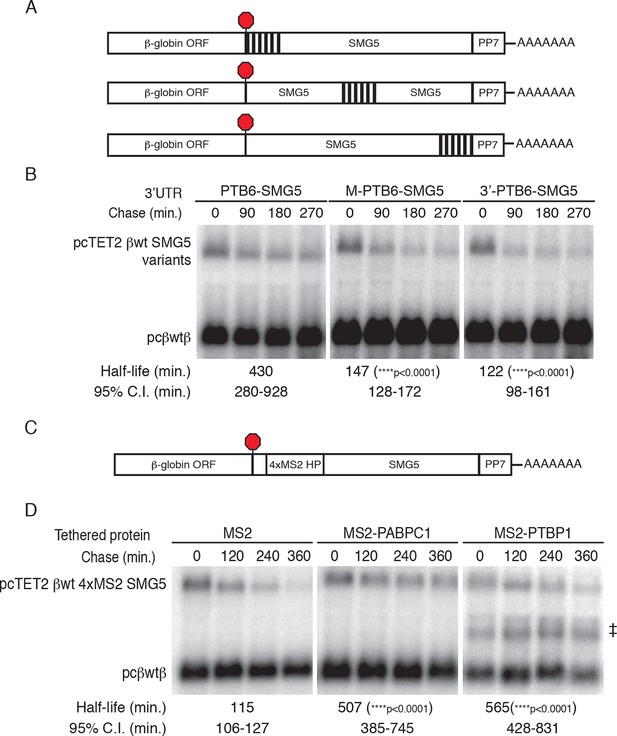
PTBP1 binding recapitulates the RSE’s position-dependent NMD inhibition.
(A) Schematic of tet-regulated β-globin reporter mRNA constructs containing six closely spaced PTBP1 binding sites at the 5’ end, in the middle, or at the 3' end of the SMG5 3’UTR. (B) Decay assays of reporter mRNAs containing the RSE at different positions in the 3'UTR. Constructs encoding the tet-regulated transcripts described in panel A (pcTET2-βwt-PTB6-SMG5, pcTET2-βwt-MPTB6-SMG5, or pcTET2-βwt-3’PTB6-SMG5; top bands) were co-transfected with the constitutively expressed wild-type β-globin reporter (pcβwtβ; bottom bands) in HeLa Tet-off cells. Remaining RNA levels at indicated time points were normalized to levels of the wild-type β-globin transfection control. Half-lives and 95% confidence intervals were obtained from 3 independent experiments (p-values from two-tailed ANCOVA analysis when compared to pcTET2-βwt-PTB6-SMG5). (C) Schematic of β-globin reporter mRNA constructs containing four MS2 coat protein binding sites at the 5’ end of the SMG5 3’UTR. (D) Tethering reporter constructs (top bands) were co-transfected with plasmids expressing the indicated MS2 fusion protein (vector control, PAPBC, or PTBP1) and the constitutively expressed wild-type β-globin reporter (pcβWTβ; bottom bands). RNA was harvested 30 min after transcription was halted by addition of doxycycline and at 2 hr intervals thereafter. Remaining RNA levels at indicated time points were normalized to levels of the wild-type β-globin transfection control. Half-lives and 95% confidence intervals were obtained from 3 independent experiments (p-values from two-tailed ANCOVA analysis when compared to pcTET2-βwt-4xMS2-SMG5 co-transfected with MS2 protein alone). ‡ denotes an aberrantly processed mRNA isoform that was excluded from quantification.
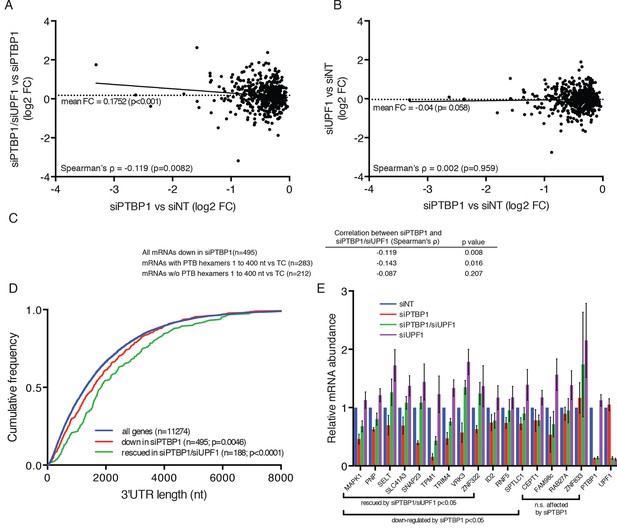
PTBP1 protects many human mRNAs with long 3’UTRs from NMD.
(A) For mRNAs from 495 genes reproducibly down-regulated by PTBP1 depletion in RNAseq of biological replicates, fold change comparing siPTBP1/siUPF1 to siPTBP1 alone was plotted vs. fold change comparing siPTBP1 to siNT. Mean fold change among all analyzed genes (dashed line; p<0.0001 in two-tailed Student’s t-test of null hypothesis of zero fold change), best fit determined by the least squares method (solid line) and correlation coefficient (Spearman’s ρ) are indicated. See also Figure 6—source data 1. (B) Mean fold change in transcript abundance upon PTBP1 depletion versus siNT treatment was plotted against the mean fold change in abundance between siUPF1 and siNT conditions for the set of mRNAs as described in A. (C) Table of Spearman’s correlation coefficients for all mRNAs reproducibly downregulated by siPTBP1, mRNAs containing one or more putative PTBP1 hexamer binding sites within 400 nt of the termination codon, and mRNAs lacking putative PTBP1 hexamers in that region. (D) mRNAs protected from NMD by PTBP1 have long 3’UTRs. Continuous distribution function (CDF) plot of annotated 3’UTR lengths among all expressed exemplar mRNAs (blue), mRNAs down-regulated by PTBP1 depletion (red), and mRNAs rescued by co-depletion of PTBP1 and UPF1 in RNA-seq analyses (green; see Materials and methods for details). Statistical significance was evaluated by two-tailed K-S test, comparing the indicated mRNA sets to the distribution of 3’UTR lengths among all exemplar mRNAs. (E) qRT-PCR analysis of mRNAs protected from NMD by PTBP1. Graph of average abundance of mRNAs normalized to housekeeping UBC control mRNAs (n=3, error bars indicate SD). Statistical significance was determined by two-tailed Student’s t-test, comparing siPTBP1 to siNT and siPTBP1/siUPF1 to siPTBP1. See also Figure 6—figure supplements 1, 2 and Figure 6—source data 1.
-
Figure 6—source data 1
Table of mRNAs down-regulated by PTBP1 depletion.
Table lists transcript accession numbers, log2 fold changes in gene expression, and p-values from one-way ANOVA analyses when comparing siPTBP1 vs. siNT and siPTBP1/siUPF1 vs. siPTBP1.
- https://doi.org/10.7554/eLife.11155.017
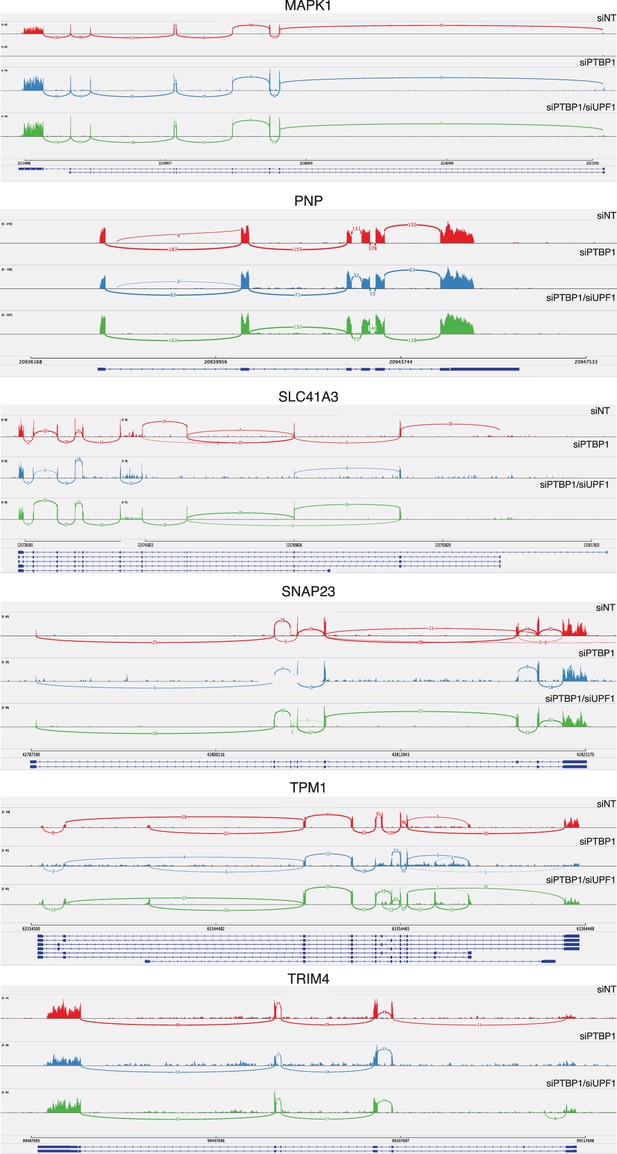
PTBP1-mediated protection does not depend on changes in splicing.
Sashimi plots were generated from reads mapping to genes rendered susceptible to NMD by PTBP1 depletion in RNA-seq and qRT-PCR experiments using Integrative Genomics Viewer software (Broad Institute). SELT, VRK3, and ZNF322 were omitted from this analysis due to insufficient read coverage spanning exons.
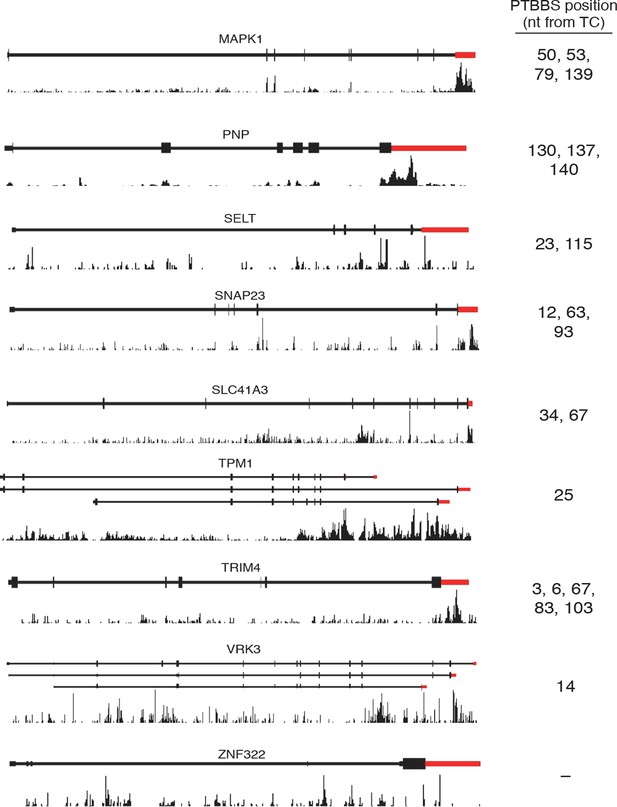
Analysis of PTBP1 binding to protected mRNAs.
Histograms of sequence reads from PTBP1 CLIP experiments (Xue et al., 2013) mapping to the indicated mRNAs were visualized using the Genomatix genome browser. Schematics of representative RefSeq transcripts are shown, and histograms are autoscaled according to maximum peak height for each gene window. Thin lines indicate introns, thick lines indicate coding regions, and red indicates 3’UTRs. Positions of putative high-affinity hexamer PTBP1 binding sites 3’ of TCs are indicated.
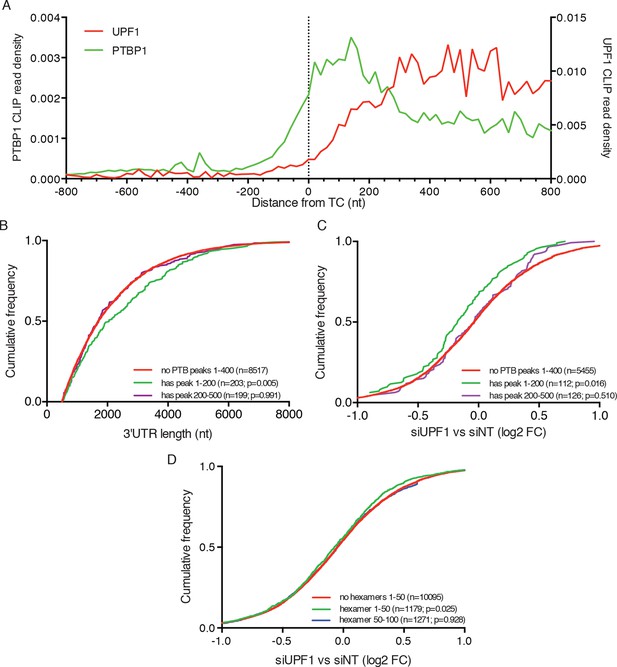
PTBP1 binding near stop codons is correlated with 3’UTR length and NMD evasion.
(A) Density of PTBP1 (green, left axis) and UPF1 (red, right axis) CLIP reads derived from peaks called using PIPE-CLIP software, plotted relative to TC position. A bin size of 20 nt was used to determine read density; bin sizes ranging from 5 nt to 20 nt and plots of peak occurrences showed similar patterns. (B) CDF plot of annotated 3’UTR lengths among mRNAs containing or lacking PTBP1 CLIP peaks centered in the indicated intervals relative to the TC. P-values were calculated in two-tailed K-S tests of 3’UTR lengths in the indicated mRNA classes compared to mRNAs lacking PTBP1 peaks. Only mRNAs with 3’UTRs greater than 500 nt were included to avoid bias due to selection of transcripts containing CLIP peaks. (C) CDF plot of log2 fold changes in mRNA abundance in UPF1 siRNA vs siNT RNAseq. mRNAs with 3’UTR lengths in the middle 50% of the overall distribution (484–2251 nt) were selected to avoid confounding effects of increased 3’UTR lengths among mRNAs with PTBP1 peaks. P-values were determined by two-tailed K-S tests, comparing the indicated mRNA classes to mRNAs lacking PTBP1 peaks from +1–400 nt. (D) CDF plot as in C. mRNAs were classified according to the presence of one or more predicted high-affinity PTBP1 hexamer binding sites in the indicated intervals relative to annotated TCs (see Materials and methods for details; the top 6 hexamers identified in previous PTBP1 CLIP seq, associated with >50% of observed CLIP peaks, were used for this analysis; Xue et al., 2009). Statistical significance was determined by two-tailed K-S tests comparing mRNAs with putative PTBP1 binding sites in the indicated positions to mRNAs lacking hexamer binding sites at positions from 1 to 50 nt downstream of the TC.
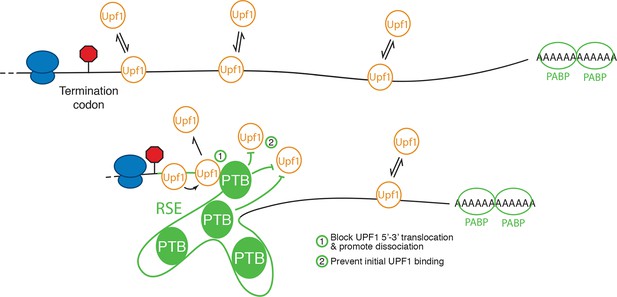
Model for NMD inhibition by PTBP1.
In the absence of the RSE or other sequences capable of recruiting PTBP1, UPF1 binds 3’UTRs in a length-dependent manner, potentiating NMD (top). PTBP1 can bind the RSE at multiple sites throughout the 400 nt sequence. This establishes an mRNP structure in the vicinity of the stop codon that is refractory to UPF1 binding, possibly by (1) blocking 5’-3’ translocation of UPF1 or (2) preventing initial binding of UPF1 to the mRNA. The NMD pathway thus judges the 3’UTR to be short and fails to degrade the mRNA.





