ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization
Figures
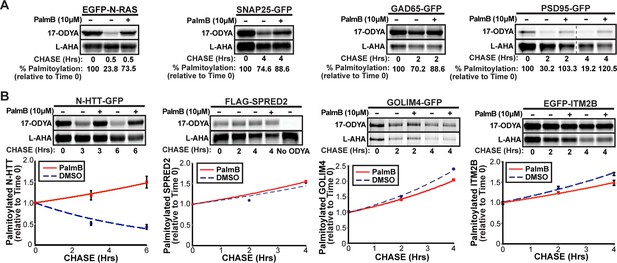
Dual-click chemistry labeling reveals differences in protein depalmitoylation dynamics.
(A) Pulse-chase analysis of established palmitoyl-proteins (N-Ras, SNAP25, GAD65, PSD95) by dual-click chemistry in the presence of DMSO (-) or 10 μM PalmB (+). Representative in-gel fluorescence scans illustrate dual detection of 17-ODYA (palmitate analogue) and L-AHA (methionine analogue) using Alexa Fluor 488 and Alexa Fluor 647, respectively. Dashed line indicates cropping of a single gel. n = 2 per substrate. (B) Pulse-chase analysis of palmitate turnover on N-HTT, SPRED2, GOLIM4, and ITM2B by dual-click chemistry as described in (A). Upper panels: representative in-gel fluorescence scans; Lower panels: Time course of substrate depalmitoyation in DMSO- and PalmB-treated cells after normalizing 17-ODYA to L-AHA signals at each chase time. n = 2, mean ± SEM. 17-ODYA, 17-octadecynoic acid; L-AHA, L-azidohomoalanine; SEM, standard error of the mean.
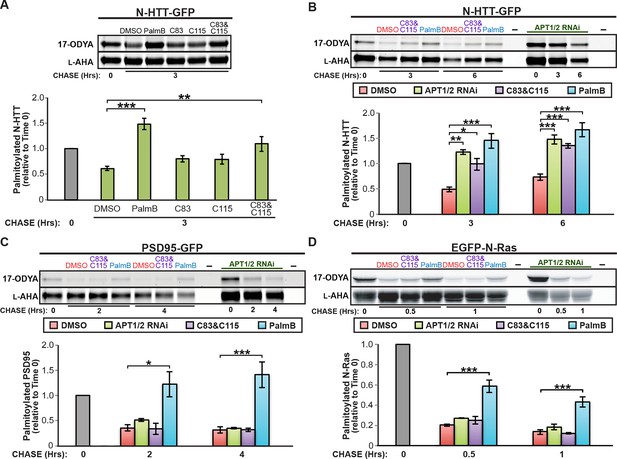
Downregulation of APT1 and APT2 inhibits HTT depalmitoylation but does not affect palmitate turnover on PSD95 or N-Ras.
(A) Pulse-chase analysis of N-HTT palmitoylation in the presence of DMSO, 10 μM PalmB, 10 μM APT1-selective inhibitor C83, and/or 10 μM APT2-selective inhibitor C115, as described in Figure 1. n = 3, mean ± SEM. (B-D) Pulse-chase analysis of (B) N-HTT, (C) PSD95, and (D) N-Ras after APT1 and APT2 knockdown (“APT1/2 RNAi”), treatment with DMSO, treatment with 10 μM C83 and 10 μM C115, or treatment with 10 μM PalmB, as described in Figure 1. n = 3, mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. SEM, standard error of the mean.
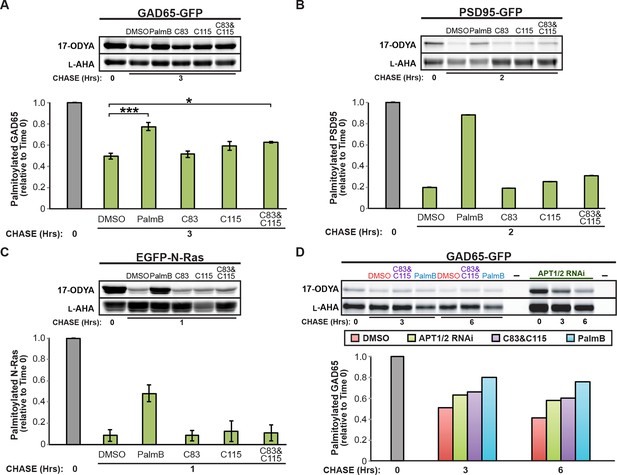
Downregulation of APT1 and APT2 inhibits GAD65 depalmitoylation but does not affect palmitate turnover on PSD95 or N-Ras.
(A-C) Pulse-chase analysis of (A) GAD65, (B) PSD95, and (C) N-Ras palmitoylation in the presence of DMSO, 10 μM PalmB, 10 μM APT1-selective inhibitor C83, and/or 10 μM APT2-selective inhibitor C115, as described in Figure 2. (D) Pulse-chase analysis of GAD65 after APT1 and APT2 knockdown (“APT1/2 RNAi”), treatment with DMSO, treatment with 10 μM C83 and 10 μM C115, or treatment with 10 μM PalmB, as described in Figure 2. *p < 0.05; ***p < 0.001.
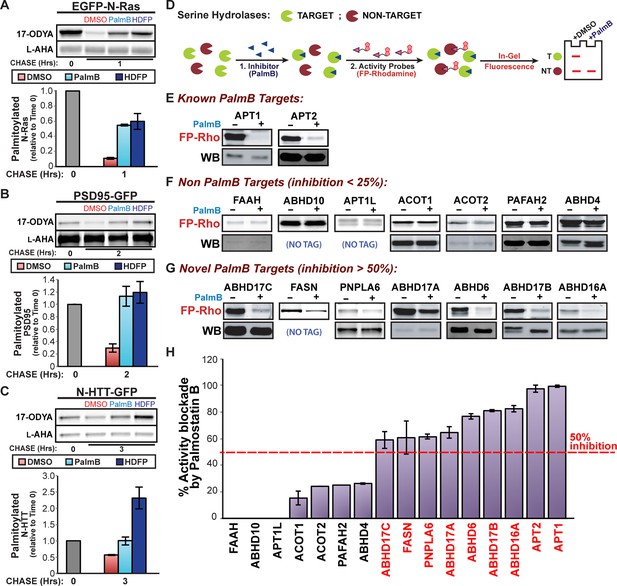
Shared targets of Palmostatin B and HDFP identified by competitive activity-based protein profiling.
(A-C) Pulse-chase analysis of (A) N-Ras, (B) PSD95, and (C) N-HTT in the presence of DMSO, 10 μM PalmB or 20 μM lipase inhibitor HDFP as described in Figure 1. n = 3 (DMSO and PalmB) or 2 (HDFP), mean ± SEM. (D) Schematic diagram of the competitive ABPP assay used in this study. (E-G) Competitive ABPP of PalmB by in-gel fluorescence (FP-Rho). 16 HDFP targets were incubated with 2 μM FP-Rho in the presence (+) or absence (-) of 10 μM PalmB. Western blots (WB) show reduced FP-Rho labeling is not due to protein loss. (H) Percent inhibition of each HDFP target by PalmB. n = 3, mean ± SEM. Candidate depalmitoylases (>50% inhibition by PalmB) are highlighted in red. SEM, standard error of the mean,
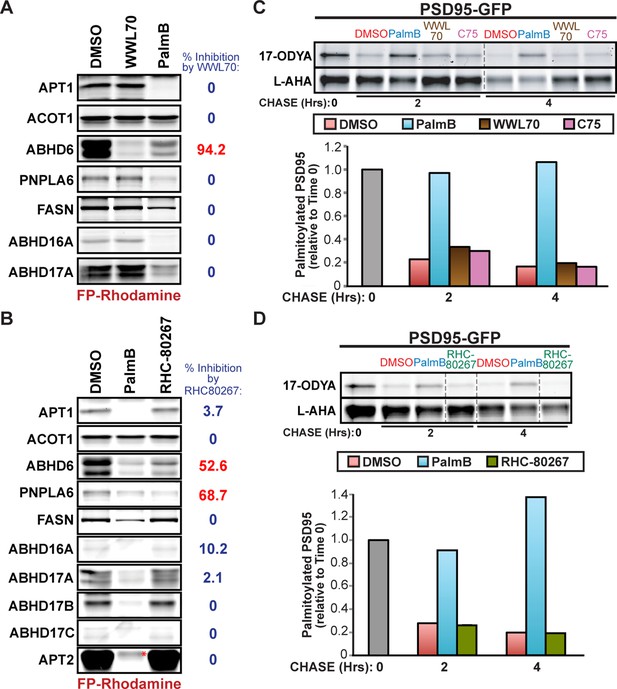
Treatment with serine hydrolase inhibitors WWL70, C75, and RHC-80267 does not affect PSD95 palmitate turnover.
(A-B) Competitive ABPP of 10 μM PalmB and (A) 10 μM WWL70 or (B) 20 μM RHC-80267 against candidate depalmitoylases and ACOT1. Percent inhibition of each enzyme is relative to DMSO. (C-D) Pulse-chase analysis of PSD95 palmitoylation in the presence of: (C) 10 μM PalmB, 10 μM WWL70, or 20 μM C75; and (D) 10 μM PalmB or 20 μM RHC-80267, as described in Figure 2. Dashed lines represent cropping of single gels. *, endogenous serine hydrolase activity unaffected by PalmB.
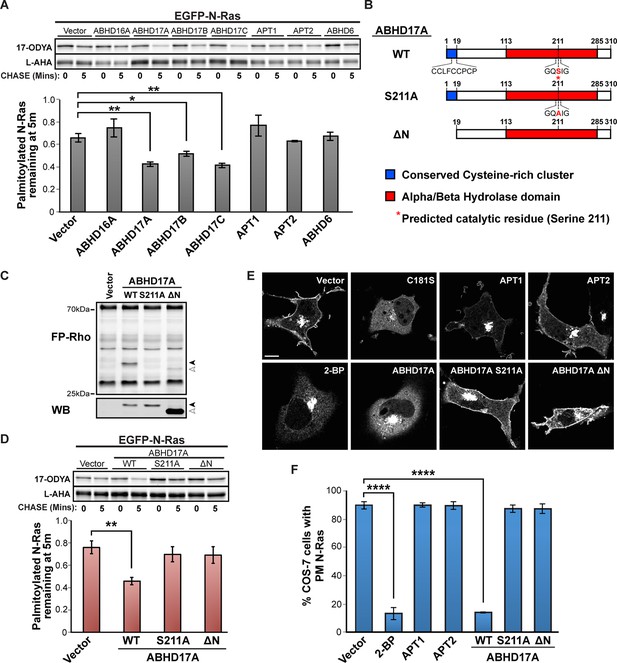
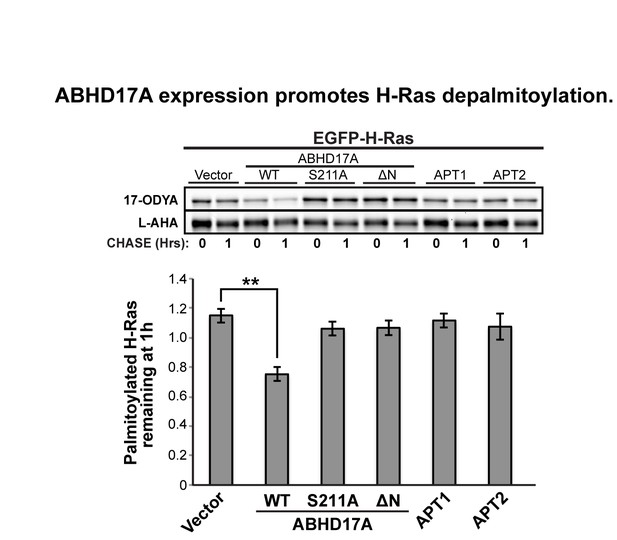
ABHD17A expression promotes N-Ras depalmitoylation and alters N-Ras subcellular localization.
(A) Pulse-chase analysis of N-Ras co-expressed with candidate mSHs as described in Figure 1. n = 3, mean ± SEM. (B) Schematic of the ABHD17A wild type, catalytically-inactive (S211A), and N-terminal truncation (ΔN) mutant proteins used in this study. (C) ABPP of ABHD17A wild type and mutant proteins by in-gel fluorescence (FP-Rho). Western blot (WB) shows proteins expressed in each condition. Filled arrowheads: ABHD17A WT and S211A; Open arrowheads: ABHD17A ΔN. (D) Pulse-chase analysis of N-Ras co-expressed with ABHD17A wild type and mutant proteins as described in Figure 1. n = 3, mean ± SEM. (E) Representative live confocal images of EGFP-N-Ras-C181S and EGFP-N-Ras localization in COS-7 cells treated with 100 μM 2-bromopalmitate (2-BP) or co-expressing the indicated thioesterases. Scale Bar = 10 μm. (F) Bar graph representing percentage of COS-7 cells with plasma membrane EGFP-N-Ras under each condition studied in (E). n = 3 (100 cells counted per trial), mean ± SEM. *p < 0.05; **p < 0.01; ****p < 0.0001. mSHs, metabolic serine hydrolases; SEM, standard error of the mean.
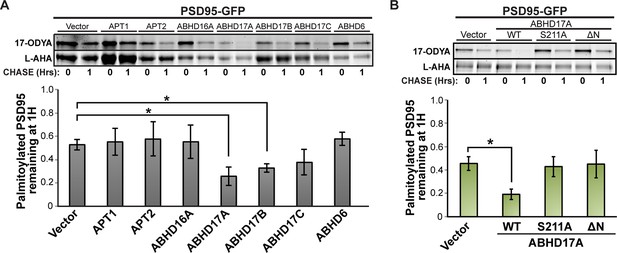
ABHD17 expression promotes PSD95 depalmitoylation.
(A) Pulse-chase analysis of PSD95 co-expressed with candidate mSHs as described in Figure 4A. n = 3, mean ± SEM. (B) Pulse-chase analysis of PSD95 co-expressed with ABHD17A wild type and mutant proteins as described in Figure 4D. n = 3, mean ± SEM. *p < 0.05. mSHs, metabolic serine hydrolases; SEM, standard error of the mean.
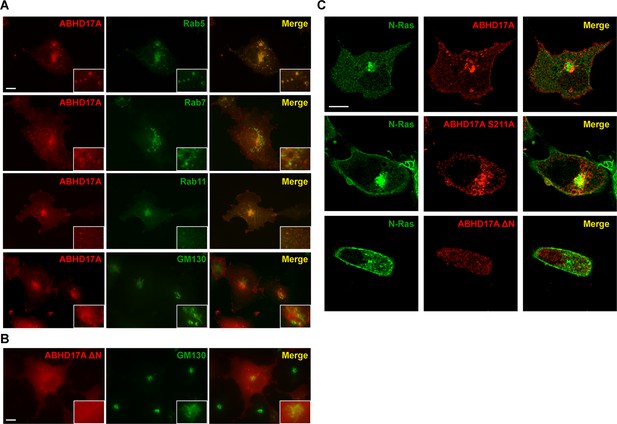
ABHD17A is localized to the plasma membrane and endosomal compartments.
(A) Localization of ABHD17A wild-type protein with markers of early endosomes (Rab5), late endosomes (Rab7), recycling endosomes (Rab11), and the Golgi apparatus (GM130) in COS-7 cells as determined by immunocytochemistry. Scale bar =10 μm. (B) Localization of ABHD17A ΔN in COS-7 cells relative to the Golgi marker GM130 by immunocytochemistry. Scale bar =10 μm. (C) Localization of mCherry-tagged ABHD17A wild type and mutant proteins co-expressed with EGFP-N-Ras in COS-7 cells by confocal microscopy. Scale bar =10 μm.
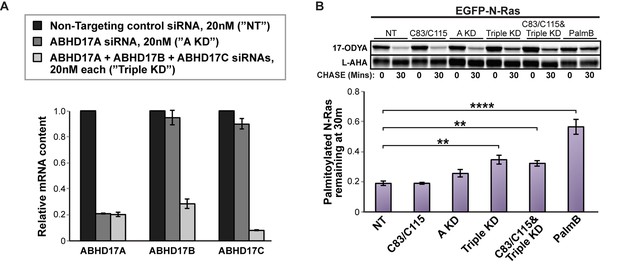
Simultaneous knockdown of ABHD17 isoforms inhibits N-Ras palmitate turnover.
(A) RT-qPCR of ABHD17A, ABHD17B, and ABHD17C transcript levels in HEK 293T cells treated with Non-Targeting siRNA (”NT”, black), ABHD17A siRNA alone (”A KD”, gray), or ABHD17A/ ABHD17B/ ABHD17C siRNAs (”Triple KD”, light gray) for 72 hr. n = 3, mean ± SEM. (B) Pulse-chase analysis of N-Ras palmitoylation in siRNA-transfected HEK 293T cells treated with vehicle (DMSO), 10 μM C83 and C115, or 10 μM PalmB as described in Figure 1. n = 3, mean ± SEM. **p < 0.01; ****p < 0.0001. SEM, standard error of the mean.
Additional files
-
Supplementary File 1
List of Metabolic serine hydrolases inhibited by HDFP.
A summary table compiling the 29 serine hydrolases targeted by HDFP (>25% activity inhibition) as determined by cABPP-SILAC (Stable isotope labeling of amino acids in culture) in (Martin et al., 2011). LYPLAL1 (APT1L) was added to this list as a candidate enzyme for Palmostatin B testing (Tian et al., 2012).
- https://doi.org/10.7554/eLife.11306.012
-
Supplementary File 2
List of cloning oligos used in this study.
A table listing PCR primers used to subclone candidate serine hydrolases for cABPP, pulse-chase/click chemistry, and confocal imaging studies.
- https://doi.org/10.7554/eLife.11306.013
-
Supplementary File 3
List of gene-specific RT-qPCR primer pairs used in this study.
A table listing gene-specific primer pairs for verification of transcript levels in HEK293T cells by RT-qPCR in Figure 5A.
- https://doi.org/10.7554/eLife.11306.014





