Genetic variation in offspring indirectly influences the quality of maternal behaviour in mice
Figures
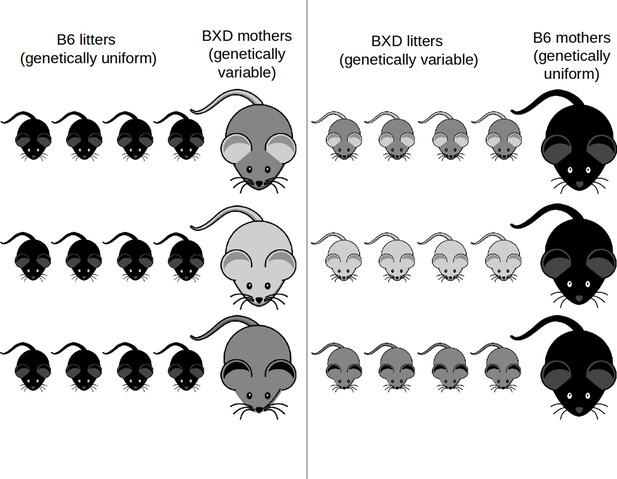
Experimental cross-foster design.
Females of different lines of the BXD strain (light to grey mice) adopt B6 offspring (dark) and B6 females (dark) adopt offspring born to females of different BXD lines. A total of 42 BXD lines with three within-line repeats plus the corresponding B6 families were set up for the experiment.
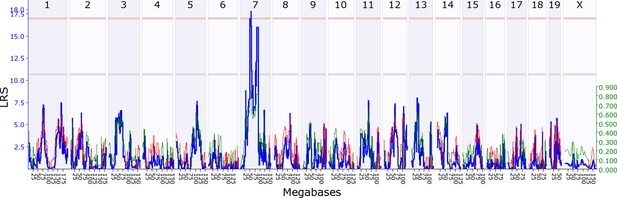
Offspring indirect genetic effect modifying maternal nestbuidling behaviour (OspIge7.1).
The figure shows an offspring genomescan of maternal nestbuilding behaviour on day 6. The blue line represents the genome scan, showing the likelihood ratio statistic (LRS) associated with each marker across the 19 autosomal and the X chromosome. The top, pink, line marks genome-wide significance, the lower, grey, line the suggestive significance threshold. The green or red line show the additive coefficient, with green showing that the DBA/2J alleles increase trait values and red that the C57BL/6J alleles increase trait values. The green axis on the right shows by how much the respective alleles increase trait values (the DBA/2J allele in offspring increases maternal nestbuilding by ~0.8).
-
Figure 2—source data 1
Source data for Figure 2 as uploaded to GeneNetwork, showing the B6 maternal nestbuilding behaviour on day 6 per pup for the BXD lines.
- https://doi.org/10.7554/eLife.11814.005
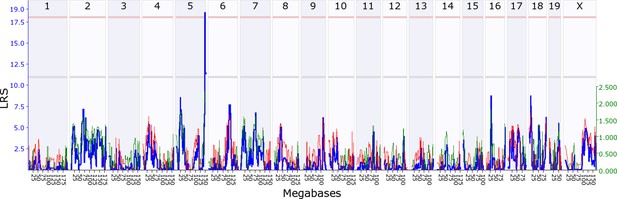
Offspring indirect genetic effect modifying maternal behaviour (OspIge5.1).
The figure shows an offspring genomescan of maternal behaviour on day 14. The blue line represents the genome scan, showing the likelihood ratio statistic (LRS) associated with each marker across the 19 autosomal and the X chromosome. The top, pink, line marks genome-wide significance, the lower, grey, line the suggestive significance threshold. The green or red line show the additive coefficient, with green showing that the DBA/2J alleles increase trait values and red that the C57BL/6J alleles increase trait values. The green axis on the right shows by how much the respective alleles increase trait values (the DBA/2J allele in offspring increases maternal behaviour by ~2.5).
-
Figure 3—source data 1
Source data for Figure 3 as uploaded to GeneNetwork, showing the B6 maternal behaviour on day 14 per pup for the BXD lines.
- https://doi.org/10.7554/eLife.11814.007
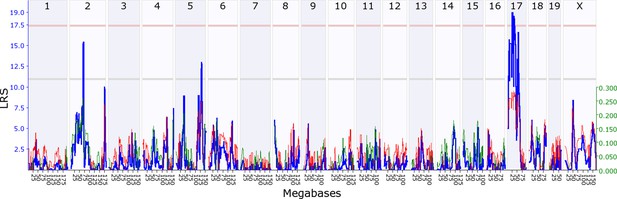
Maternal indirect genetic effect modifying offspring growth (MatIge17).
The figure shows a maternal genomescan of offspring growth in the second postnatal week. The blue line represents the genome scan, showing the likelihood ratio statistic (LRS) associated with each marker across the 19 autosomal and the X chromosome. The top, pink, line marks genome-wide significance, the lower, grey, line the suggestive significance threshold. The green or red line show the additive coefficient, with green showing that the DBA/2J alleles increase trait values and red that the C57BL/6J alleles increase trait values. The green axis on the right shows by how much the respective alleles increase trait values (the C57BL/6J allele in mothers increases offspring growth by ~0.25).
-
Figure 4—source data 1
Source data for Figure 4 as uploaded to GeneNetwork, showing the B6 offspring growth in the second postnatal week per pup for the BXD lines.
- https://doi.org/10.7554/eLife.11814.009
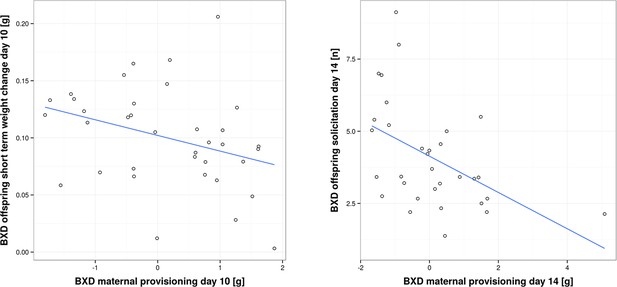
Correlation between offspring and maternal traits in biological BXD families.
The first panel shows the correlation between BXD offspring short-term weight change per pup and provisioning of their corresponding biological BXD mother on day 10 per pup. The second panel shows the correlation between the level of BXD offspring solicitation per pup on day 14 and their mother’s provisioning per pup.
-
Figure 5—source data 1
Source data for Figure 5 as uploaded to GeneNetwork, showing BXD offspring short-term weight change on day 10 per pup, BXD maternal provisioning on day 10, BXD offspring solicitation on day 14 per pup and BXD maternal provisioning on day 14 for the BXD lines.
- https://doi.org/10.7554/eLife.11814.012
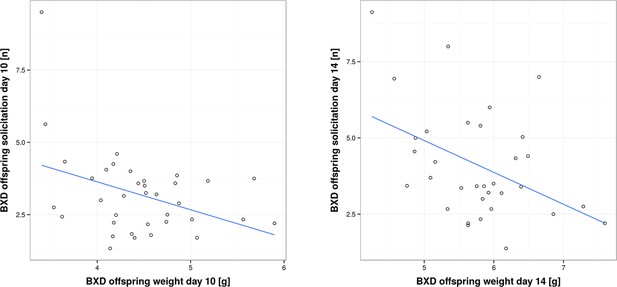
Correlation between per pup offspring solicitation and corresponding body weight in BXD lines on day 10 and day 14, respectively.
https://doi.org/10.7554/eLife.11814.013-
Figure 6—source data 1
Source data for Figure 5 as uploaded to GeneNetwork, showing BXD offspring solicitation on day 10 per pup, BXD offspring weight on day 10 per pup, BXD offspring solicitation on day 14 per pup and BXD offspring weight on day 14 per pup for the BXD lines.
- https://doi.org/10.7554/eLife.11814.014
Tables
Summary of direct and indirect genetic effects. We list the loci, followed by their position in Mb, the 1.5 LOD confidence interval (Dupuis and Siegmund, 1999), the genome-wide peak marker LRS and LOD score and associated p-value and the number of genes within the interval.
| Loci | QTL position | Confidence interval | Max LRS | Max LOD | Max P | Number of genes |
|---|---|---|---|---|---|---|
| MatDge1.1 | 168.32 | 165.24 - 172.06 | 17.03 | 3.70 | 0.069 | 68 |
| OspDge5.1 | 23.827 | 17.82 - 24.62 | 17.85 | 3.88 | 0.046 | 73 |
| OspIge5.1 | 146.68 | 145.20 - 147.65 | 18.65 | 4.05 | 0.038 | 40 |
| OspIge7.1 | 53.68 | 47.76 - 56.65 | 17.85 | 3.88 | 0.039 | 232 |
| 81.49 | 76.12 - 90.92 | 16.06 | 3.49 | 0.087 | 133 | |
| MatDge10.1 | 19.09 | 18.61 - 21.83 | 22.37 | 4.86 | 0.008 | 30 |
| MatIge17.1 | 23.32 | 11.48 - 31.17 | 19.02 | 4.13 | 0.022 | 422 |
| 33.02 | 31.32 - 40.65 | 18.57 | 4.03 | 0.028 | 321 |
Additional files
-
Supplementary file 1
A) Functional and further details about the genes within the OspIge5.1 QTL for B6 maternal behaviour on day 14, obtained from GeneNetwork, Entrez genes, and Mouse Genome Informatics. Potential candidate genes in this region are Cyp3a16, Cyp3a44, Cyp3a11, Cyp3a25 and Cyp3a41a because of their involvement in steroid hormone biosynthesis. B) Functional and further details about the genes within the MatDge1.1 QTL for BXD nestbuilding on day 6, obtained from GeneNetwork, Entrez genes, and Mouse Genome Informatics. Potential candidate genes in this region include: Hsd17b7, as it is involved in steroid hormone synthesis, and hormonal regulation is needed to initiate nestbuilding behaviour (Gammie et al., 2007; Keisala et al., 2007; Bester-Meredith and Marler, 2012); Pou2f1, Lmx1a and Rgs4 as they are linked to activity related phenotypes; finally, Pbx1 and Ddr2 have been linked to craniofacial morphology, which may affect the ability to make nests (Schneider et al., 2012). C) Functional and further details about the genes within the MatDge10.1 QTL for BXD maternal behaviour on day 6, obtained from GeneNetwork, Entrez genes, and Mouse Genome Informatics. A potential candidate gene is Ifngr1 as it is related to depression-like behaviour, which in turn has been linked to reduced maternal care (Smith et al., 2004; Malkesman et al., 2008). D) Functional and further details about the genes within the OspDge5.1 QTL for BXD offspring solicitation on day 6, obtained from GeneNetwork, Entrez genes, and Mouse Genome Informatics. Cdk5 might be a good candidate as it is involved in several neuronal annotations (e.g. axonogenesis and synaptic transmission), and mutants have no suckling reflex (Ohshima et al., 1996).
- https://doi.org/10.7554/eLife.11814.015
-
Supplementary file 2
Details of correlation analyses.
- https://doi.org/10.7554/eLife.11814.016





