The E3 ubiquitin ligase ZNRF2 is a substrate of mTORC1 and regulates its activation by amino acids
Figures
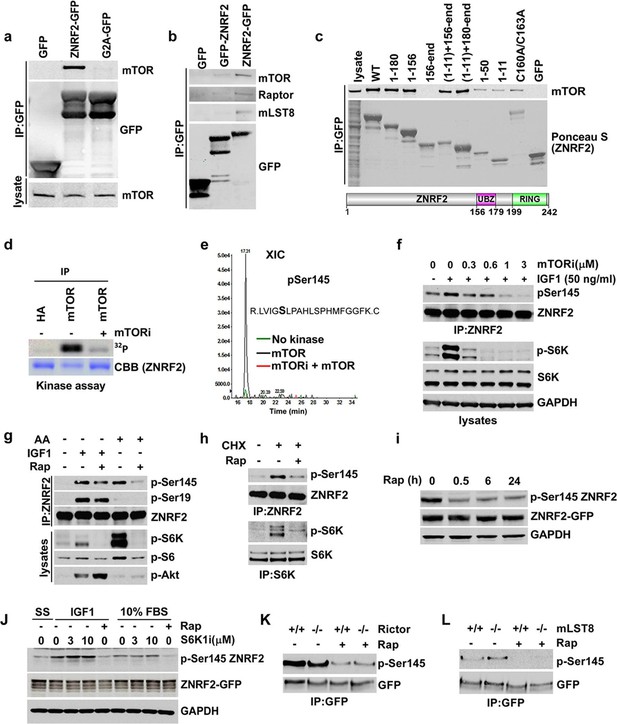
ZNRF2 binds to mTOR and is a substrate of mTORC1.
(a) Lysates of cells expressing GFP control, ZNRF2-GFP and a myristoylation mutant G2A-GFP (with GFP-tag at the C-terminus) were subject to immunoprecipitation with GFP-Trap beads. Precipitates were immunoblotted with the indicated antibodies. (b) HEK293 Flp-In cells that stably express GFP or GFP-tagged (N-terminus and C-terminus) ZNRF2 were harvested in 0.3% CHAPS lysis buffer and GFP tagged proteins were immunoprecipitated with GFP-Trap beads. Precipitates were analyzed by Western blotting with the indicated antibodies. (c) Full-length ZNRF2 and the indicated fragments of ZNRF2 (all with C-terminal GFP tags) were tested for binding to endogenous mTOR. C160A/C163A represents ZNRF2 with the zinc-coordinating cysteines of the UBZ domain changed to alanines. Below, schematic diagram of full-length ZNRF2 containing UBZ (zinc finger) and Ring (catalytic) domains is shown. (d) Kinase assays were performed using mTOR immunopurified from HEK293 cells and bacterially purified GST-ZNRF2 as substrate in the presence of Mg2+[γ-32P]ATP for 30 min. Where indicated, 1 µM of catalytic mTOR inhibitor (Ku-0063794) was added to the reaction and incubated for 10 min at 4oC before the addition of the GST-ZNRF2. Reactions were subjected to SDS-PAGE and autoradiography. CBB, Coommassie brilliant blue. (e) As in (d), except that phosphorylated GST-ZNRF2 was digested with trypsin and tryptic peptides were analyzed by LC-MS on an ABI 4000 Q-TRAP system using precursor ion scanning in negative mode, searching for the (PO3-) ion (-79 Da). The extracted ion chromatograph for phospho-Ser145 is presented. (f) HEK293 cells stimulated with IGF1, in the presence or absence of the indicated amounts of catalytic mTOR inhibitor (Ku-0063794), were tested for phosphorylation of S145 by Western blotting after immunoprecipitating endogenous ZNRF2. The phosphorylation status of Thr389 p70S6-kinase (p-S6K) was assayed. p-S6K is a marker of mTORC1 activation. S6K and GAPDH were used as controls. (g) HEK293 cells were starved of amino acids (1.5hr) and stimulated with IGF1 or amino acids, in the presence or absence of rapamycin. Endogenous ZNRF2 was immunoprecipitated and its phosphorylation (p-S19 and p-S145) was assayed by Western blotting. (h) HEK293 cells were starved of amino acids as in (g) and cycloheximide was added in the presence or absence of rapamycin. Phosphorylation of ZNRF2 was assayed as in (g). (i) Cell lysates of HEK293 Flp-In cells stably expressing ZNRF2-GFP were treated with 20 nM rapamycin for the indicated times. Phosphorylation of ZNRF2 was assayed as in (g). (j) HEK293 Flp-In cells that stably express ZNRF2-GFP were stimulated with IGF1 or serum in the presence or absence of the indicated amounts of S6K1 inhibitor (PF-4708671). Cell lysates were tested for phosphorylation of S145 of ZNRF2. (k–l) Rictor and mLST8 wild-type and knock-out MEFs were transfected with ZNRF2-GFP for 36 hr. Cells were treated with 20 nM rapamycin for 30 min in fresh media containing 10% FBS. ZNRF2-GFP was immunoprecipitated and precipitates were immunoblotted with p-S145 antibody of ZNRF2.
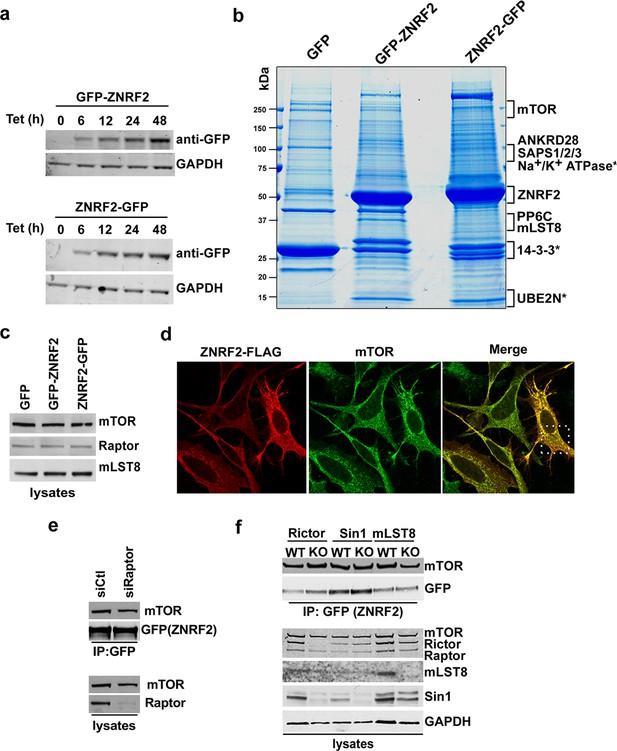
ZNRF2 is a target of mTORC1 and interacts with mTOR.
(a) Plasmids pcDNA5.1–FRT–TO (Invitrogen), expressing ZNRF2 with a N-terminal GFP tag (top) or a C-terminal GFP tag (bottom), were stably integrated into HEK293 Flp-In T-Rex cells. The expression of proteins was induced with tetracycline (1 µg/ml) for the indicated times. (b) HEK293 Flp-In T-Rex cells that stably express GFP (control) and GFP-tagged (N-terminus and C-terminus) ZNRF2 were induced with tetracycline (1 µg/ml) for 36 hr before lysis. These extracts were subjected to immunoprecipitation with GFP-Trap beads and precipitates were subjected to SDS-PAGE. The gel was fixed and stained with Colloidal Blue. The gel lanes were cut into slices, as indicated, and the proteins were digested with trypsin before mass spectrometric fingerprinting. (c) Input blots of Figure 1b. (d) HeLa cells were transfected with ZNRF2-FLAG for 12 hr. Images of cells co-immunostained for FLAG (red) and mTOR (green) are shown, together with the merged images. Inset shows a higher magnification of a selected field. (e) HEK293 Flp-In cells stably expressing ZNRF2-GFP were transfected with siRNAs targeting raptor. Cell extracts were subjected to immunoprecipitation with GFP-Trap and analyzed by Western blotting with the antibodies indicated. (f) Rictor, Sin1 and mLST8 wild-type and knockout MEFs were transfected with ZNRF2-GFP for 48 hr. Cells were lysed in 1% Triton lysis buffer. ZNRF2-GFP was immunoprecipitated with GFP-Trap® beads and the binding to mTOR was tested.
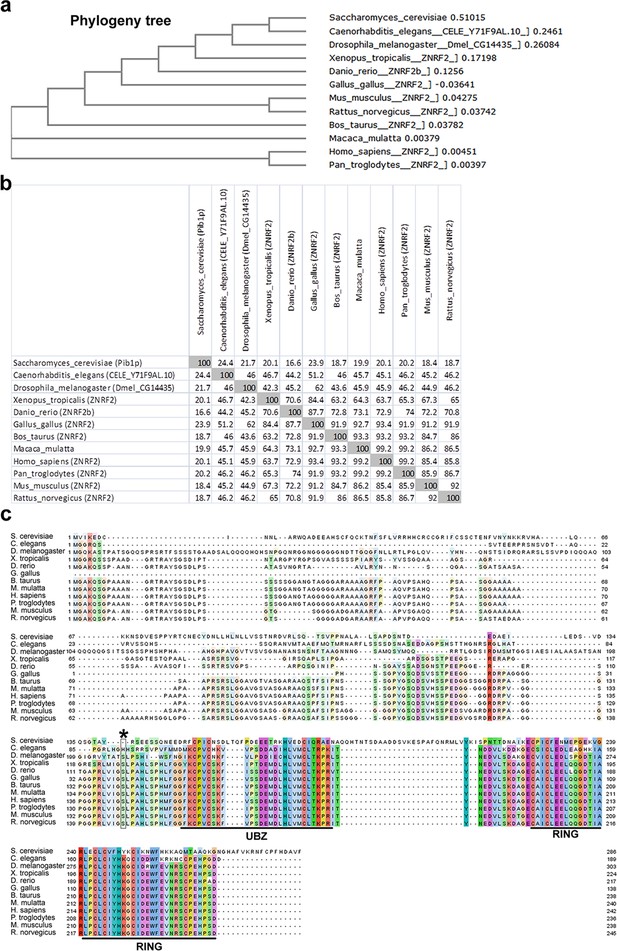
Conservation of ZNRF2 in eukaryotes.
(a) Phylogeny tree depicting conservation of amino acid sequence of ZNRF2 orthologues among eukaryotes. (b) Matrix plot showing percentage amino acid identity among ZNRF2 orthologues. (c) Amino acid sequence alignment showing conservation of N-myristoylation signal, Ser145 residue (asterisk), UBZ and RING domains. Putative ZNRF2 orthologue in D. melanogaster is an uncharacterized protein called CG14435, which shows 73% amino acid identity with human ZNRF2, with an E-value of 10–49 and sequence coverage of 40%. The putative ZNRF2 orthologue in C. elegans is an uncharacterized protein called Y71F9AL.10, which shows 56% amino acid identity with human ZNRF2, with an E-value of 6x10-42 and sequence coverage of 48%. The putative ZNRF2 orthologue in S. cerevisiae is a protein called Pib1p, which shows 47% amino acid identity with human ZNRF2, with an E-value of 2x10-6 and sequence coverage of 14%, according to BLAST alignment. Importantly, putative ZNRF2 orthologues in vertebrates, D. melanogaster, C. elegans and S. cerevisiae share the highest similarity with human ZNRF2 in the UBZ and RING domains. Notably, the fruit fly and nematode orthologues of ZNRF2 also have a conserved N-myristoylation signal.
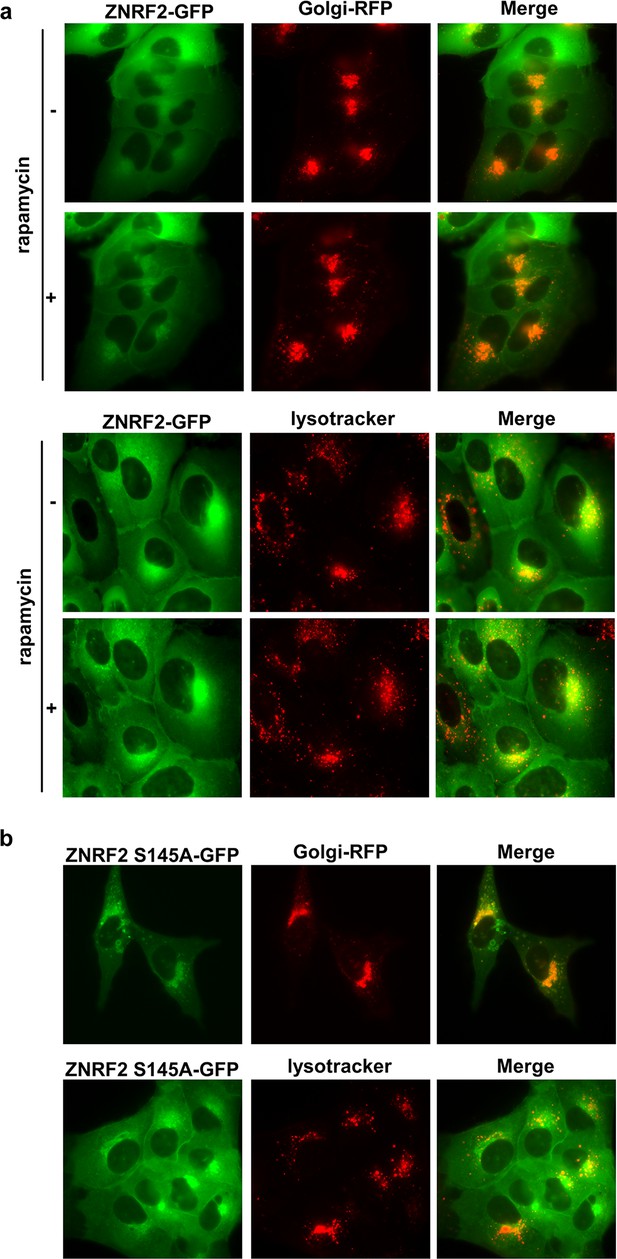
Ser145 phosphorylation of ZNRF2 promotes its dissociation from membranes.
(a) Live cells from U2OS-Flp-In expressing stably ZNRF2-GFP were imaged under serum conditions in the presence or absence of 200 nM rapamycin. The colocalization with Golgi marker (CellLight Golgi-RFP) and lysosome marker (Lysotracker), was tested. (b) Live cells from ZNRF2-Ser145A U2OS-Flp-In stables were imaged under serum conditions. Colocalization with Golgi marker (CellLight Golgi-RFP) and lysosome marker (lysotracker) is shown.
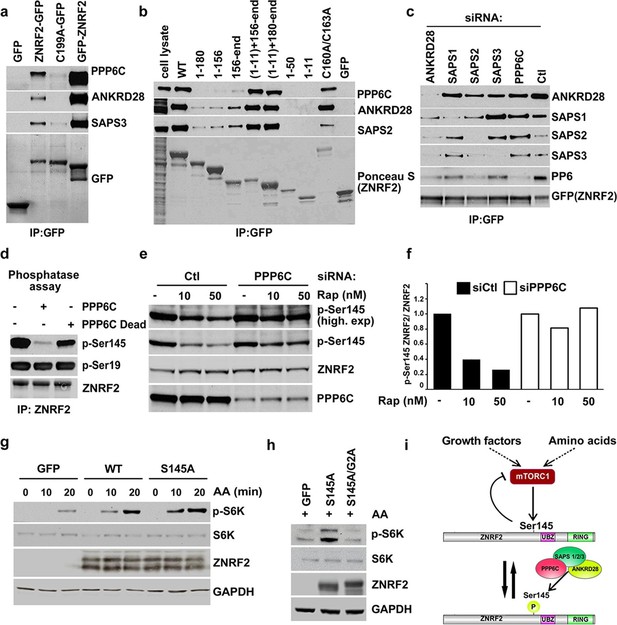
PP6 interacts with ZNRF2 and dephosphorylates phosphoS145 of ZNRF2.
(a) Lysates of cells expressing GFP, GFP-ZNRF2 and ZNRF2-GFP (with GFP-tag at N- and C-terminus, respectively), and ligase-dead C199A-ZNRF2-GFP were subject to immunoprecipitation with GFP-Trap beads. Precipitates were immunoblotted with the indicated antibodies against the PP6 complex components. (b) As in Figure 1c, except that the immunoprecipitates were blotted for the PP6 complex component. (c) HEK293 Flp-In cells stably expressing ZNRF2-GFP were transfected with siRNAs targeting individual PP6 components or control (Ctl). Cell extracts were subjected to immunoprecipitation with GFP-Trap and analyzed by Western blotting with the antibodies indicated. (d) Purified ZNRF2-GFP was incubated with wild-type or catalytically-inactive (H55Q/R85A) recombinant PPP6C-FLAG for 2 hr and then analyzed for phosphorylation of p-S145 and p-S19 of ZNRF2. (e) HEK293 Flp-In cells expressing ZNRF2-GFP were transfected with control (Ctl) siRNA or siRNA targeting PPP6C and were incubated with two different concentrations of rapamycin (10 and 50 nM) to inhibit mTORC1. p-S145 and total ZNRF2 were assessed. (f) The ratio of p-S145 ZNRF2/ZNRF2 of Figure 2e is presented. (g) HEK293 cells were transfected to express GFP (control) and untagged ZNRF2 (wild-type and S145A mutant) were starved of amino acids (1.5hr), and stimulated with amino acids for 10 or 20 min. Western blotting was used to measure the phosphorylation status of S6K and levels of S6K and ZNRF2. GAPDH was used as the loading control. (h) HEK293 transfected with GFP control or S145A mutant or a combination of S145A with myristoylation mutant G2A were starved of amino acids and stimulated with amino acids for 20 min. Cell lysates were immunoblotted as in (g). (i) Model depicting ZNRF2 which is phosphorylated on Ser145 by mTORC1 and this site is dephosphorylated by PP6. Phosphorylation of Ser145 contributes to the release of ZNRF2 from membranes into cytosol.
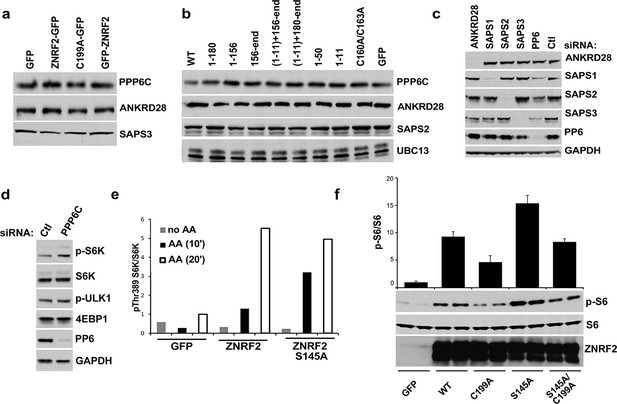
PP6 interacts with and dephosphorylate ZNRF2.
(a) Input blots of Figure 2a. (b) Input blots of Figure 2b. (c) Input blots of Figure 2c. (d) HEK293 cells transfected with control siRNA (Ctl) or siRNA targeting PP6 were serum starved. Cell lysates were analyzed for p-S6K (Th389), p-ULK1 (S757), 4EBP1, S6K, PP6 and GAPDH. (e) p-S6K/S6K ratio of immunoblots in Figure 2f, representative of 3 independent experiments. (f) HEK293 transfected with GFP control, WT, E3 ligase dead mutant (C199A), S145A mutant or a combination of S145A with E3 ligase dead mutant (S145A/C199A) were starved of amino acids and stimulated with amino acids for 20 min. Cell lysates were immunoblotted with the indicated proteins from biological duplicates. The ratio of p-S6/S6 of immunoblots is presented in the graph.
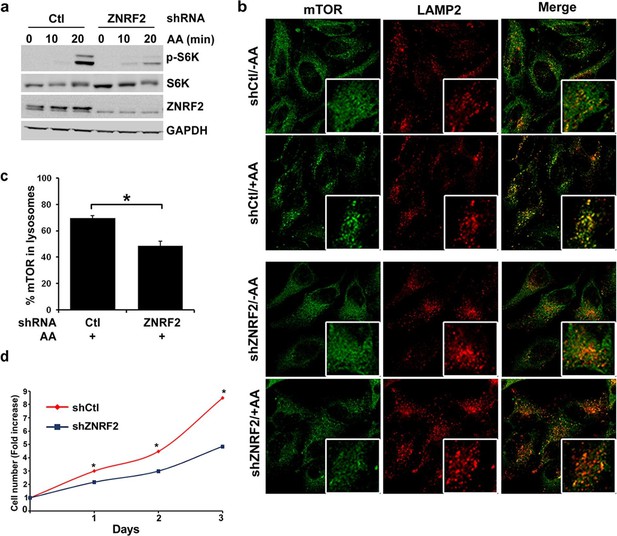
Depletion of ZNRF2 attenuates the activation of mTORC1 by amino acids .
(a) HeLa cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 were starved of amino acids (1.5 hr), and stimulated with amino acids for 10 or 20 min. Cell lysates were subjected to immunoblotting with the indicated antibodies. p-S6K is a marker of mTORC1 activation. (b) HeLa cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 were starved of amino acids (-AA), and stimulated with amino acids for 20 min. Images of HeLa cells (with or without amino acids) co-immunostained for mTOR (green) and LAMP2 (red) are shown, together with the merged images. Inset shows a higher magnification of a selected field. (c) The graph displays the quantification of cells displaying lysosomal spots of mTOR fluorescence after addition of amino acids. N = ~100 cells per condition. Data are presented as mean ± S.E.M from two independent experiments. Two-tailed Student’s t tests were used for the pairwise comparison. *p < 0.0001. (d) Cell viability of HeLa cells after knockdown of ZNRF2 was assessed daily using CellTiter-Glo Luminescent Assay. Data are mean ± S.E.M of biological triplicates from two independent experiments.
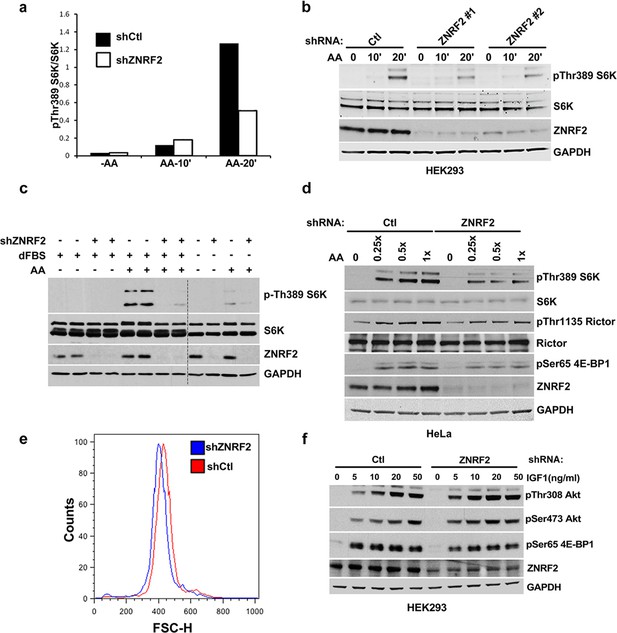
Depletion of ZNRF2 decrease the activation of mTORC1 by amino acids.
(a) The p-S6K/S6K ratio of immunoblots in Figure 3a. (b) HEK293 cells expressing control shRNA (Ctl) or two independent shRNAs targeting ZNRF2 were starved of amino acids for 1.5 hr, and stimulated with amino acids for 10 or 20 min. Western blotting was used to measure the phosphorylation status and levels of the indicated proteins. (c) HEK293 cells expressing control shRNA (Ctl) and shRNAs targeting ZNRF2 were starved of amino acids in the presence of 10% dialyzed FBS for 1.5 hr and stimulated with amino acids for 15 min. Western blotting was used to measure the phosphorylation status of S6K and levels of the indicated proteins. (d) HeLa cells expressing control shRNA (Ctl) and shRNAs targeting ZNRF2 were amino acids starved for 1.5 hr, and stimulated with the indicated concentrations of amino acids for 20 min. Western blotting was used to measure the phosphorylation status and levels of the indicated proteins. (e) Cell size of HeLa cells was measured after ZNRF2 knockdown using flow cytometry. The x-axis shows Forward Scatter (FSC), which reflects cell size. The counts are shown on the y-axis. Data shown are representative of two independent experiments performed in duplicate. (f) HEK293 cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 were serum starved (15hr) and stimulated with the indicated amount of IGF1 for 20 min. Lysates were immunoblotted with the indicated antibodies.
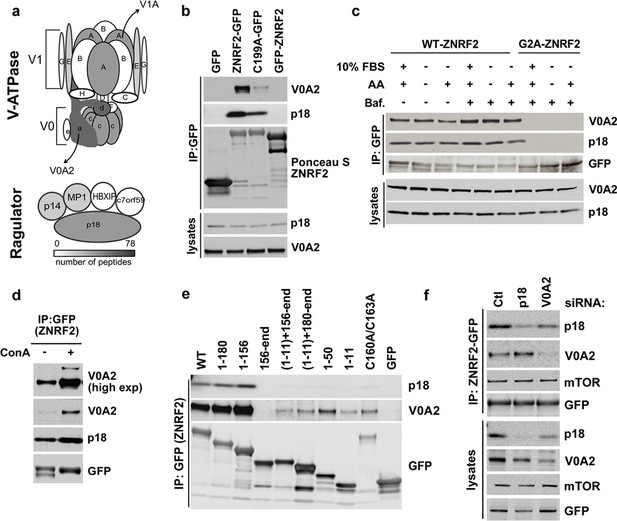
ZNRF2 interacts with V-ATPase and Ragulator.
(a) Cartoon representation of V-ATPase and Ragulator subunits from mass spectrometry analyses of immunoprecipitates of ZNRF2-GFP overexpressed in HEK293 cells. The subunits are colour-coded according to the number of peptides identified by mass spectrometry and a scale is shown at the bottom. V0 and V1 subunits of the V-ATPase are presented in small or capital letters, respectively. (b) Lysates of cells expressing GFP, GFP-ZNRF2, ZNRF2-GFP (with GFP-tag at N- and C-terminus, respectively), or ligase-dead C199A-ZNRF2-GFP were subject to immunoprecipitation with GFP-Trap beads. The immunoprecipitates were immunoblotted with antibodies against the V0A2 component of V-ATPase and the p18 component of Ragulator. (c) HEK293 Flp-In cells that stably express ZNRF2-GFP or G2A-ZNRF2-GFP were treated with 2 µM bafilomycin for 60 min in the presence of serum or amino acid free medium, or amino acid free medium with 15 min stimulation with amino acids. The lysates were subjected to immunoprecipitation with GFP-Trap beads and the precipitates were blotted with V0A2 subunit of V-ATPase and p18. (d) HEK293 Flp-In cells that stably express ZNRF2-GFP were treated with 1 µM concanamycin A (ConA) for 90 min. ZNRF2–GFP was immunoprecipitated and the interaction with V0A2 subunit of V-ATPase and p18 was tested. (e) Full-length ZNRF2 and the indicated fragments of ZNRF2 (all with C-terminal GFP tags) were tested for binding to endogenous V0A2 (V-ATPase component) and p18 subunit of Ragulator. (f) HEK293 Flp-In cells stably expressing ZNRF2-GFP were transfected with siRNA targeting control (Ctl), p18 or V0A2. The lysates were subjected to immunoprecipitation with GFP-Trap beads. Precipitates were immunoblotted with the indicated antibodies.
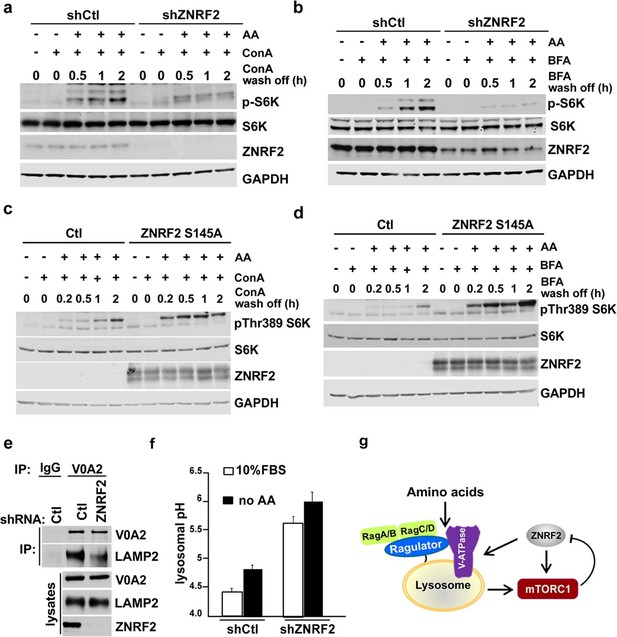
Knockdown of ZNRF2 affects V-ATPase function and lysosomal pH.
(a) HEK293 cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 were starved of amino acids in the presence of 1 µM concanamycin A (conA) for 90 min. Then, ConA containing media was washed off and cells were stimulated with amino acids for the indicated times. Cell lysates were immunoblotted with the indicated antibodies. (b) As in (a) except that cells were incubated with brefeldin A (BFA) instead of ConA. (c) HEK293 cells transfected with untagged ZNRF2 Ser145Ala or mock (Ctl) and were treated as in (a). (d) HEK293 cells transfected with untagged ZNRF2 Ser145Ala or mock (Ctl) and were treated as in (b). (e) HeLa cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 were subjected to lysis in hypotonic buffer with chemical crosslinker DSP as described in Materials and methods. After immunoprecipitation with IgG (control) or V0A2 antibody, the LAMP2 containing compartments were assessed by immunoblotting with the indicated antibodies. (f) HEK293 cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 were incubated for overnight with dextran-Oregon Green 514. Prior to pH measurements, the cell culture media was replaced with fresh media containing serum or amino acid-free media for 2 hr. The 490/440 fluorescence ratios were plotted as a function of pH and fitted to a Boltzmann sigmoid curve. Data are mean ± S.D. of biological triplicates and are representative of three independent experiments. (g) Model depicting the positive influence of ZNRF2 on V-ATPase, and hence amino acid activation of mTORC1. Also shown is the negative feedback in which mTORC1 phosphorylates and inhibits ZNRF2.
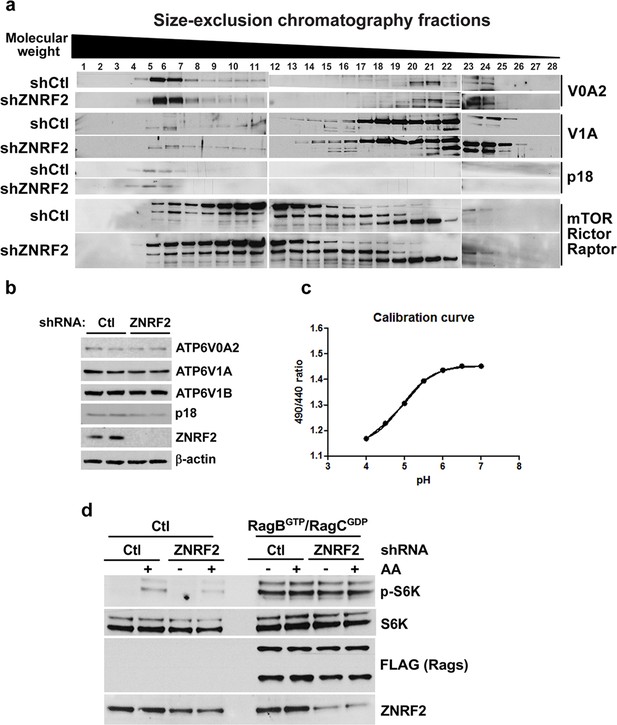
Knockdown of ZNRF2 affects V-ATPase function and lysosomal pH.
(a) HEK293 cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 were harvested in 0.3% CHAPS lysis buffer and 1.2 mg of each sample was injected onto a Superose 610/300GL column and analysed by size-exclusion chromatography. Fractions of 500 µl were collected, diluted 1:1 in sample buffer, and subject to immunoblotting with the indicated antibodies. (b) HEK293 cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 grown under 10% FBS were immunoblotted for the indicated V-ATPase subunits and Ragulator protein levels. (c) A calibration curve used for Figure 5f was created by loading HEK293 cells with Dextran-Oregon Green, washing out excess dye and resuspending the cells in K+ isotonic buffers at different pH values. The 490/440 fluorescence ratios were plotted as a function of pH and fitted to a Boltzmann sigmoid curve. (d) HEK293 cells expressing control shRNA (Ctl) or shRNAs targeting ZNRF2 were transfected with FLAG only vector (control) or RagBGTP/RagCGDP-FLAG tagged proteins for 36 hr. The cells were starved of amino acids, and stimulated for 20 min with amino acids. The cell lysates were immunoblotted as indicated.
Additional files
-
Supplementary file 1
V-ATPase and Ragulator subunits identified in ZNRF2-GFP immunoprecipitates.
- https://doi.org/10.7554/eLife.12278.014





