The Sec7 N-terminal regulatory domains facilitate membrane-proximal activation of the Arf1 GTPase
Figures
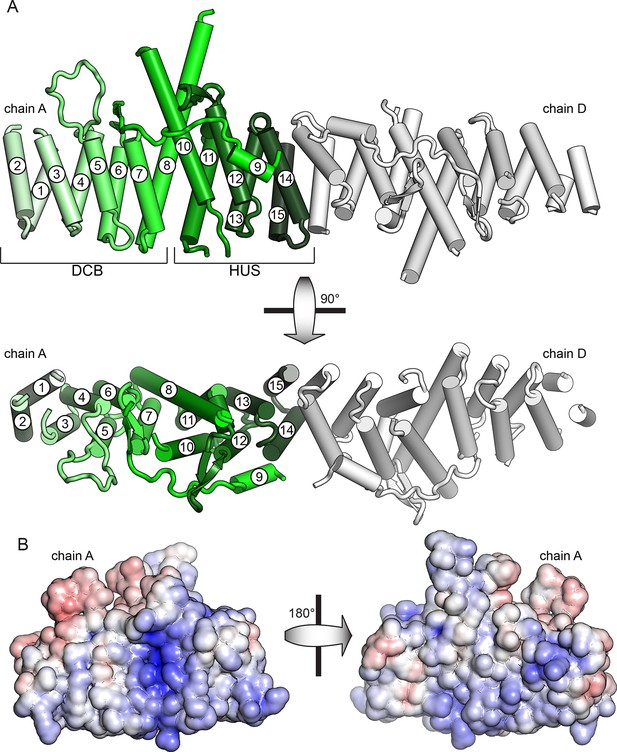
Crystal structure of T. terrestris Sec7 DCB/HUS domain (residues 1–458)
(A) Chains A (green) and D (white) are shown; chain A is colored light to dark N to C, and helices are numbered N to C. The entire asymmetric unit is shown in supplement 1. Electron density is shown in supplement 2. The DCB/HUS interface is shown in supplement 3. (B) The charge potential surface of chain A alone, as calculated by APBS (Baker et al., 2001; Dolinsky et al., 2007), is colored on a red-white-blue gradient from -10 kT/e to +10 kT/e.
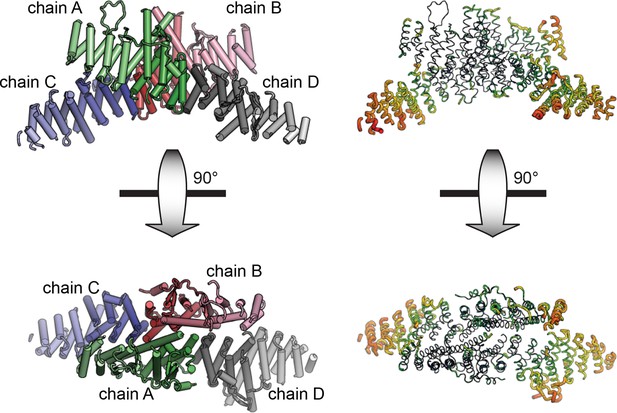
Asymmetric unit of crystal structure.
The entire asymmetric unit is shown on the left, with each chain colored light to dark N to C. A=green, B=red, C=blue, D=gray. The asymmetric unit is shown in B-factor putty form on the right (thicker chain indicates larger relative atomic B-factors).
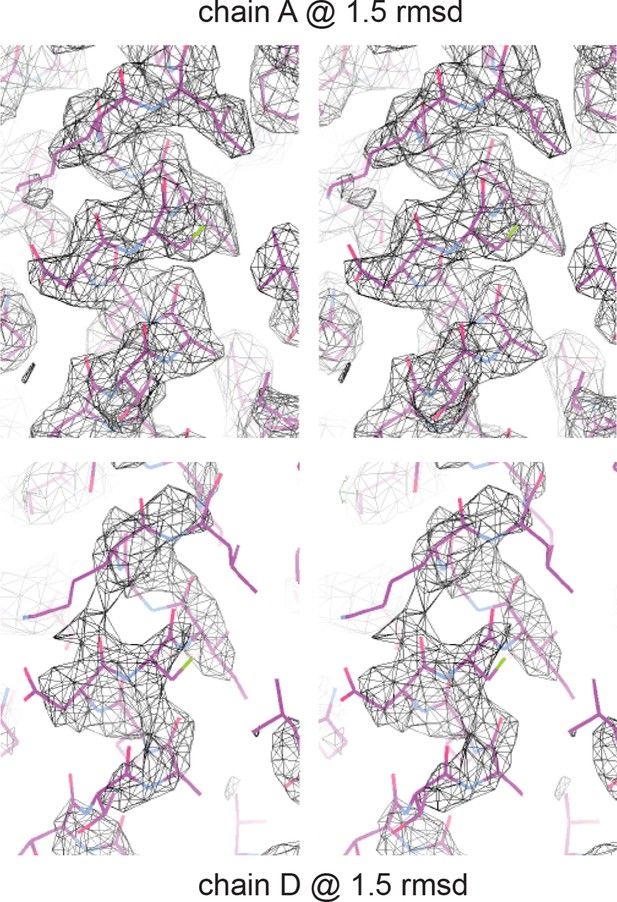
Electron density of crystal structure.
Weighted composite omit maps of helix 4 including residues D297, K301, and F305 are shown. Chain A, above, was used for structural analysis; chain D, below, was modeled on the basis of chain A.
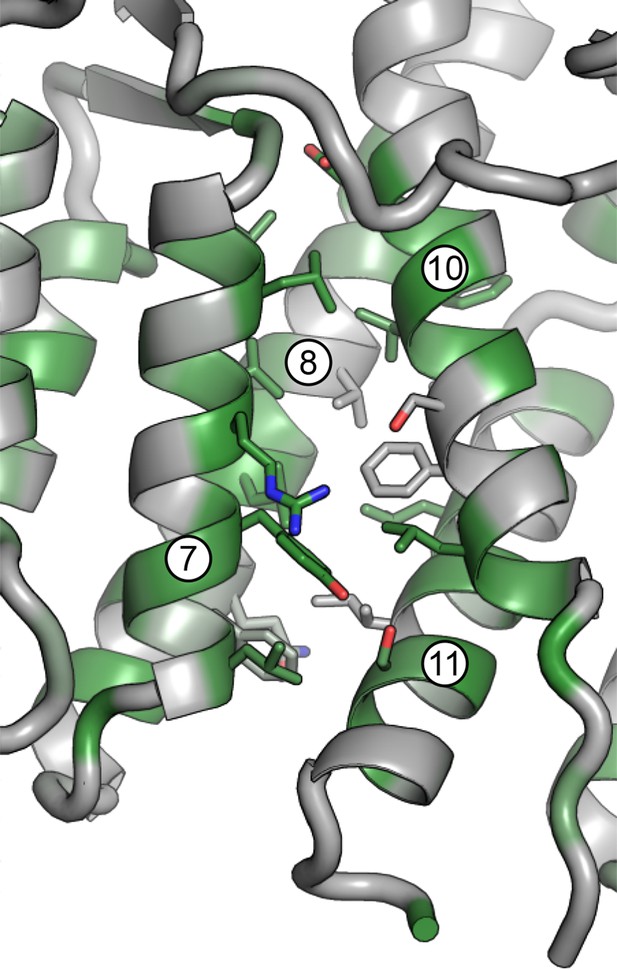
Magnified view of the DCB/HUS interface.
Packing of helices 7 and 8 (DCB) and 10 and 11 (HUS) is shown, including participating side chains. Conserved residues are colored in green.
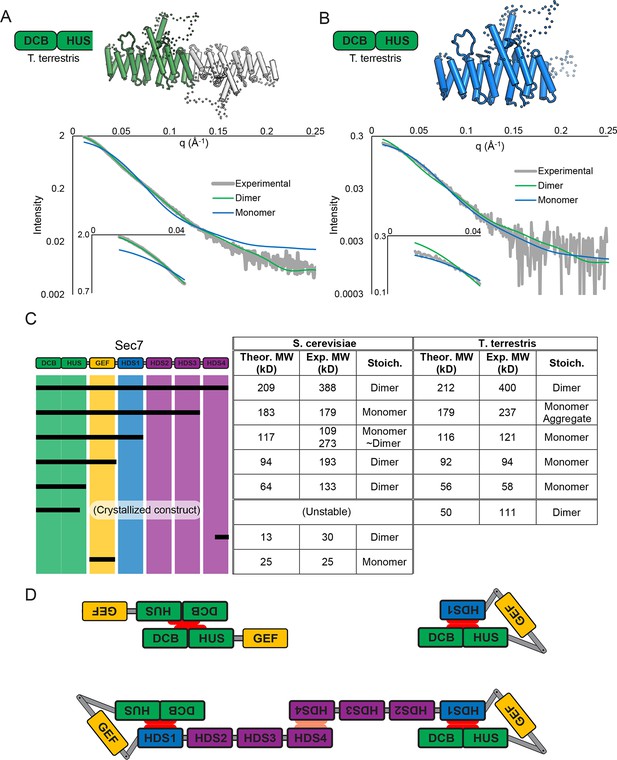
Sec7 dimerizes primarily via the HDS 4 domain.
(A) CORAL (Petoukhov et al., 2012) was used to fit the T. terrestris Sec7 DCB/HUS domain structure (residues 1–458) to SAXS data collected on the same construct, accounting for the presence of unresolved loops modelled as dotted lines. For comparison, a similar calculation using only a single chain is shown with a significantly worse fit. (B) A single monomer from (A), fixing the previously modeled loops in place and adding additional residues at the C-terminus, was fit to SAXS data collected on T. terrestris Sec7(1–492); as the added region is expected to comprise an alpha helix in addition to a connecting loop, BUNCH was used in place of CORAL for modeling. A similar fit of the dimeric form is shown for comparison. (C) The solution molecular weights of the indicated S. cerevisiae and T. terrestris constructs were determined by MALS. Comparison to the predicted monomeric mass based on primary sequence yields the stoichiometry. SAXS results from S. cerevisiae Sec7ΔC are shown in supplement 1. Results from corresponding S. cerevisiae Gea2 constructs are shown in supplement 2. Original MALS traces of all S. cerevisiae and T. terrestris constructs are shown in supplement 3. (D) Schematic model of S. cerevisiae cis and trans interactions of truncated constructs. Red represents a hypothesized interface between HDS1 and DCB/HUS, the latter half of which can self-stabilize by dimerization in the absence of HDS1. Orange represents the HDS4 dimerization interface. T. terrestris is hypothesized to possess a divergent DCB/HUS interaction interface with less need of stabilization by dimerization.
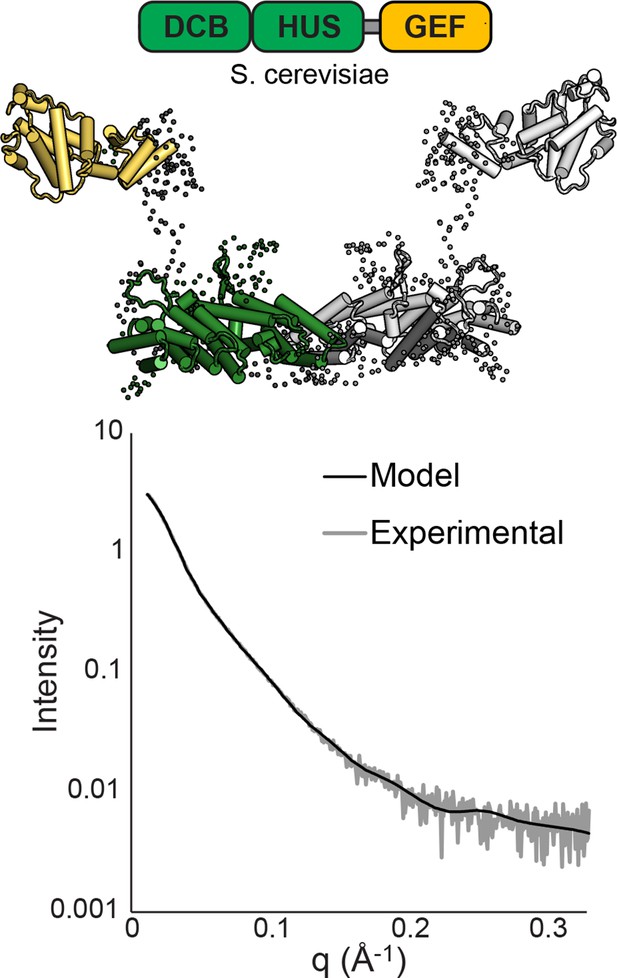
SAXS analysis of S. cerevisiae Sec7ΔC.
BUNCH (Petoukhov and Svergun, 2005) and CRYSOL (Svergun et al., 1995) were used to model T. terrestris Sec7 DCB/HUS domain (residues 1–458), S. cerevisiae Gea2 GEF domain (residues 570–714), and appropriate linker residues to SAXS data collected on S. cerevisiae Sec7∆C (residues 203–1017) with P2 symmetry. While too many degrees of freedom exist to interpret the resulting model as an accurate structural assembly, the placement of the GEF domain at a distance from the DCB/HUS domain in order to fit the data suggests a flexible linkage.
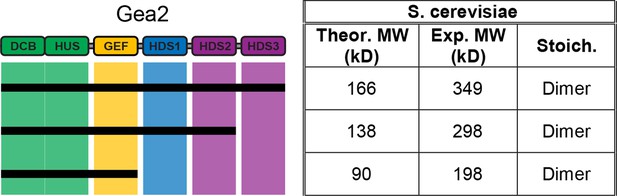
MALS analysis of S. cerevisiae Gea2.
Solution molecular weights of the indicated S. cerevisiae Gea2 constructs were determined by MALS.
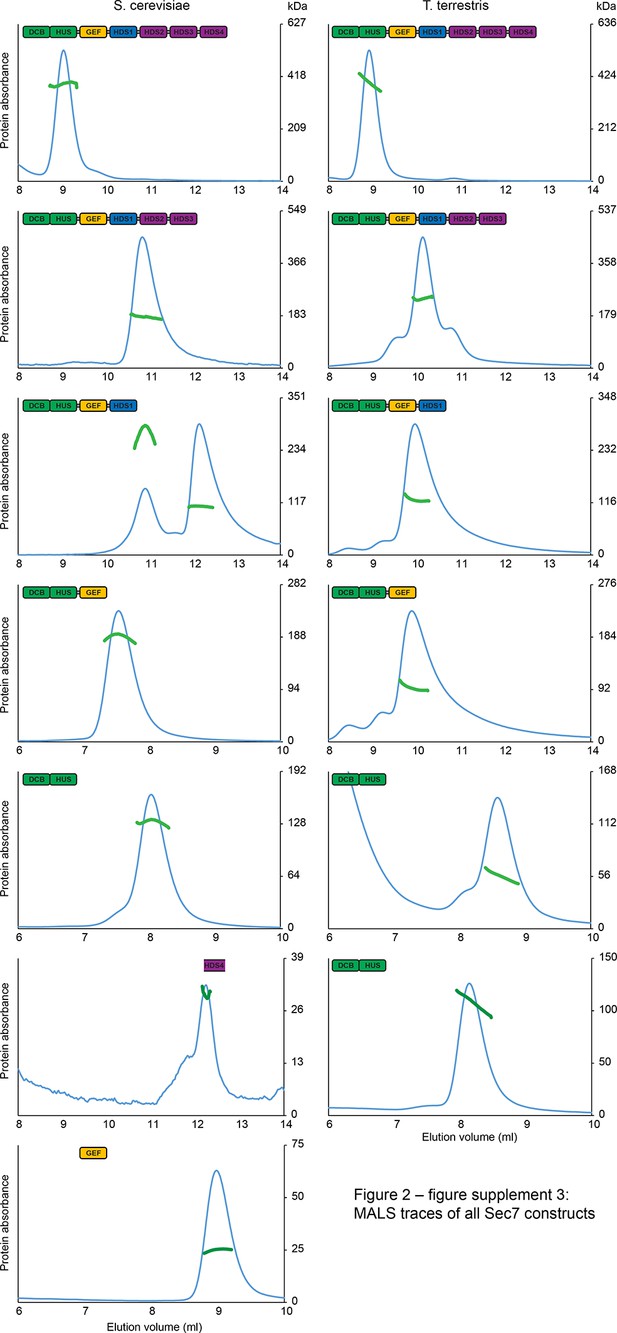
MALS traces of S. cerevisiae and T. terrestris Sec7 constructs.
Normalized gel filtration peak profiles of all Sec7 constructs are shown with MALS molecular weight measurements superimposed. The right-hand molecular weight axis for each panel is scaled to each construct’s calculated mass, with monomeric, dimeric, and trimeric masses indicated. Due to the wide variation in construct size, a single gel filtration column was not used for every construct, leading to the variation in elution profiles seen.
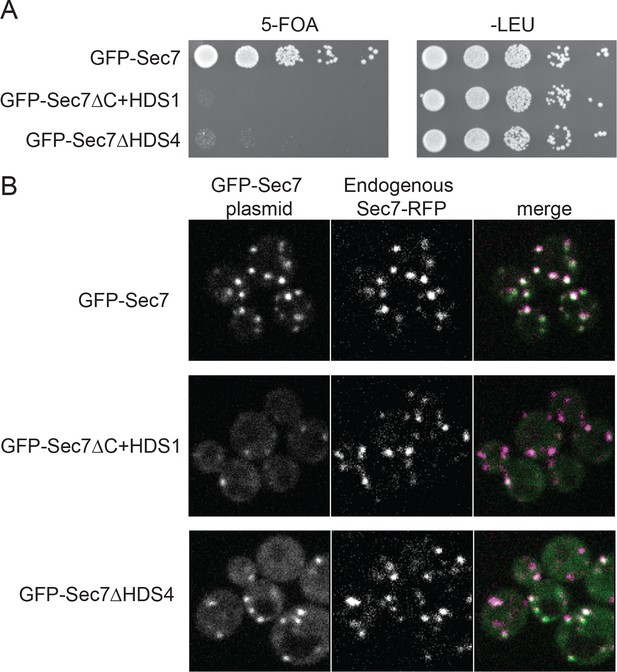
The HDS4 domain of Sec7 is important for function but dispensable for TGN localization.
(A) Centromeric plasmids encoding GFP-Sec7 constructs expressed from the SEC7 promoter were introduced into a SEC7 plasmid shuffling strain (CFY409). Growth on 5-FOA measures the ability of the construct to complement the sec7∆ mutation. (B) The same plasmids were imaged in an otherwise wild-type strain expressing endogenously tagged Sec7-RFPMars (‘Sec7-RFP’) (CFY578).
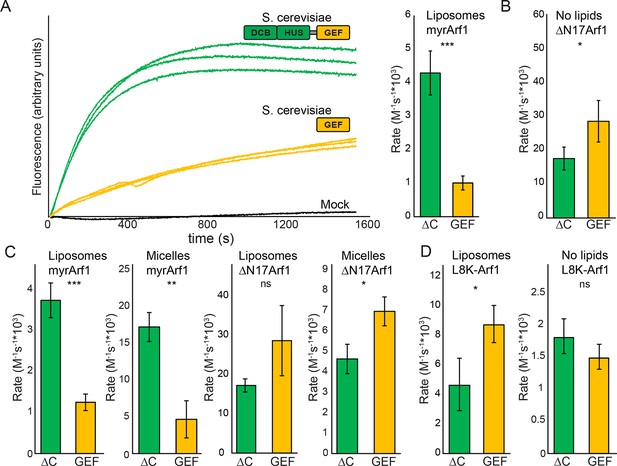
Stimulation of GEF activity by the DCB/HUS domain depends on the presence of lipids.
(A) Triplicate nucleotide exchange curves of S. cerevisiae Sec7ΔC (green traces) or isolated GEF domain constructs (yellow traces) acting on myristoylated Arf1 substrate in the presence of liposomes are shown against a mock exchange reaction (black trace) (left). The curves were fit to a single exponential and normalized to Sec7 concentration to extract the exchange reaction rates (right). Error bars denote 95% confidence intervals, n=3. Purity of all constructs is demonstrated in supplement 1. These measurements of exchange by tryptophan fluorescence are compared to complementary measurements of Arf1 membrane binding in supplement 2. (B) Exchange reaction rates of S. cerevisiae Sec7 constructs acting on the ΔN17Arf1 substrate in the absence of lipids. GEF activity of T. terrestris constructs is shown in supplement 3. GEF activity following Arl1 preincubation is shown in supplement 6, with corresponding physical interaction analysis in supplements 4 and 5. (C) Parallel reactions of S. cerevisiae Sec7 constructs acting on myristoylated Arf1 and ΔN17Arf1 in the presence of liposomes and micelles. (D) Nucleotide exchange by S. cerevisiae Sec7 constructs on non-myristoylated L8K-Arf1 in the presence and absence of liposomes.
Purity of constructs used for kinetic assays.
2.5 μg of each construct used for biochemical assays were separated by SDS-PAGE and stained by Coomassie to assess purity.
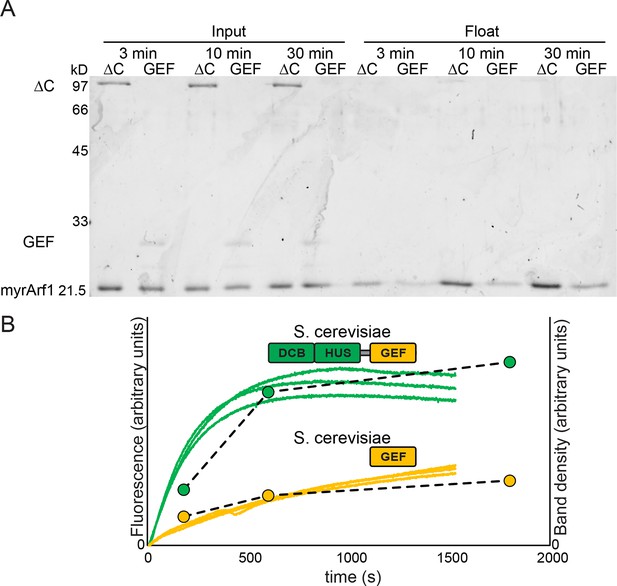
Measurement of Sec7 GEF kinetics by liposome flotation.
2 μM myristoylated Arf1-GDP was incubated with liposomes, excess GMPPNP, and. 67 μM Sec7ΔC or GEF domain for the indicated amount of time and put on ice to stop the reaction. Lipid-bound protein (i.e. activated Arf1) was assayed by liposome flotation as described previously (Richardson et al., 2012). (A) Input fractions and float/lipid-bound fractions were analyzed by SDS-PAGE and colloidal Coomassie; float fraction load amounts were normalized to the recovered lipid fraction on the basis of fluorescence of DiR dye included in the liposomes. Note that band staining intensity correlates with molecular weight, and the GEF domain construct is ~¼ the mass of the Sec7ΔC construct. (B) Quantified band densities (circles and dashed lines) are overlaid on the reaction kinetics measured by fluorescence in Figure 4A (solid lines).
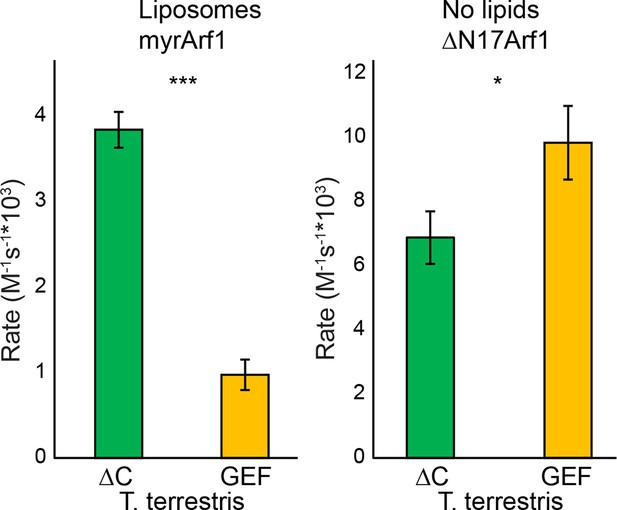
GEF activity of T. terrestris Sec7.
T. terrestris Sec7ΔC and isolated GEF constructs were assayed for rate of nucleotide exchange of myristoylated Arf1 in the presence of liposomes (left) and ΔN17Arf1 in the absence of liposomes (right).
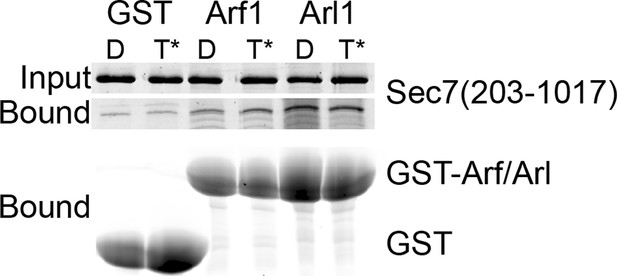
GST pulldown analysis of Arf1 and Arl1 interaction with Sec7ΔC.
Stable interaction of Arf1 and Arl1 with S. cerevisiae Sec7ΔC was assayed by pulldown of Sec7 with a great excess of GST-GTPase bound to GDP (denoted D) or GMPPNP (denoted T*). Following SDS-PAGE, protein was visualized by Coomassie staining. There was no apparent nucleotide-dependent interaction observed between Sec7ΔC and Arf1-GTP or Arl1-GTP in this experiment.
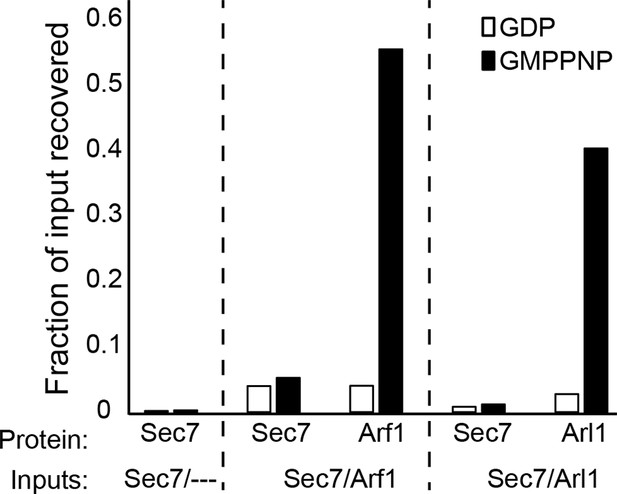
Liposome pelleting analysis of Arf1 and Arl1 interaction with Sec7ΔC.
Stable interaction of membrane-bound Arf1 and Arl1 with S. cerevisiae Sec7ΔC (as a potential effector) was assayed in the context of membranes by liposome pelleting as described previously (Paczkowski et al., 2012) following EDTA-mediated loading with the indicated nucleotide. The fraction recovered is calculated relative to a baseline of liposome-free pelleting. There was no apparent interaction observed between Sec7ΔC and membrane-bound Arf1-GTP or Arl1-GTP in this experiment.
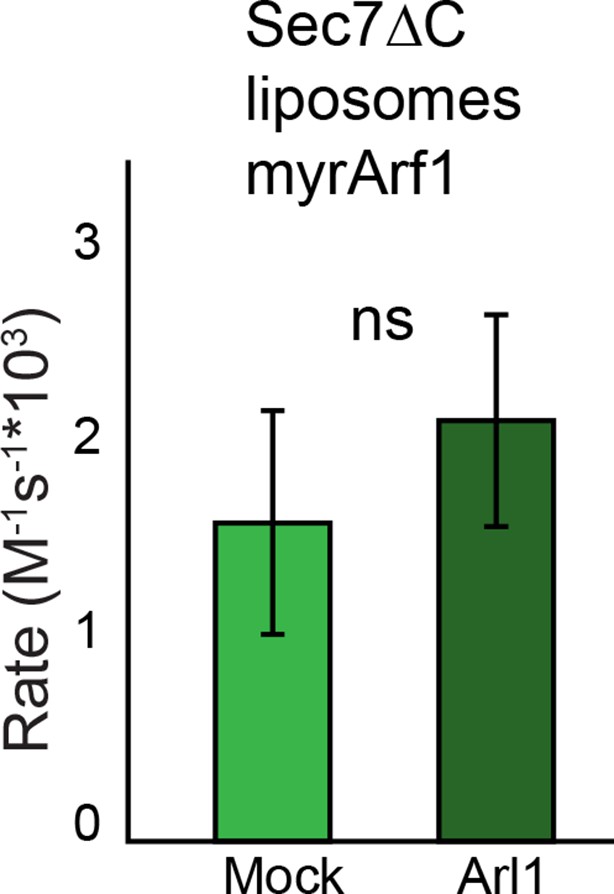
Effect of Arl1 preincubation on Sec7 GEF activity.
S. cerevisiae Sec7ΔC was assayed for rate of nucleotide exchange of Arf1 in the presence of liposomes and Arl1-GMPPNP as described previously (McDonold and Fromme, 2014).
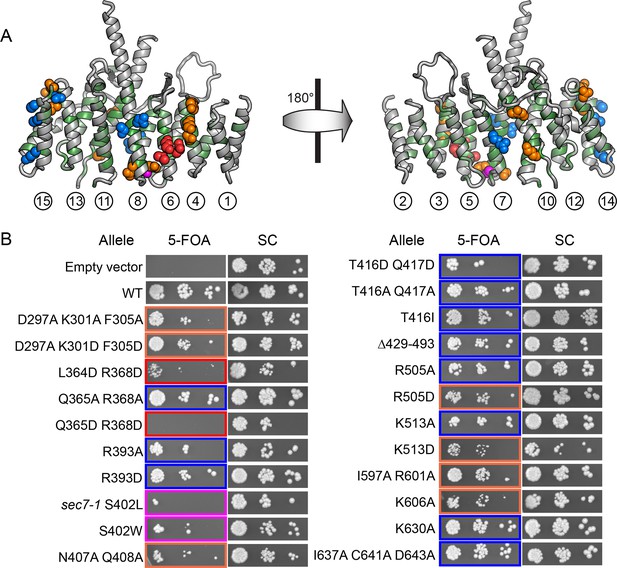
Conserved DCB/HUS surface regions mediate Sec7 function.
(A) Locations of all S. cerevisiae mutants tested are shown as space-filling spheres mapped on the T. terrestris structure, with backbone colored by conservation. Positions of mutations resulting in stronger temperature sensitive growth phenotypes are colored red (e.g., S. cerevisiae Q365/R368), positions with weaker phenotypes are colored orange (e.g., S. cerevisiae D297/K301/F305), and positions with no growth phenotype are colored blue; the position corresponding to the sec7-1 mutation (S402L) is colored magenta. (B) Using a plasmid-shuffling assay, CEN plasmids bearing GFP-tagged Sec7 or Sec7f with the indicated mutations expressed via their endogenous promoter were tested for their ability to rescue a genomic sec7 deletion in a sensitized arf1Δ/ARF2 strain (CFY863). Growth of serial 10-fold dilutions after 3 days at 37°C is shown, comparing the shuffled strains on 5-FOA to their parents growing in parallel on synthetic complete media (SC). Changes in the number or size of colonies indicates a growth defect.
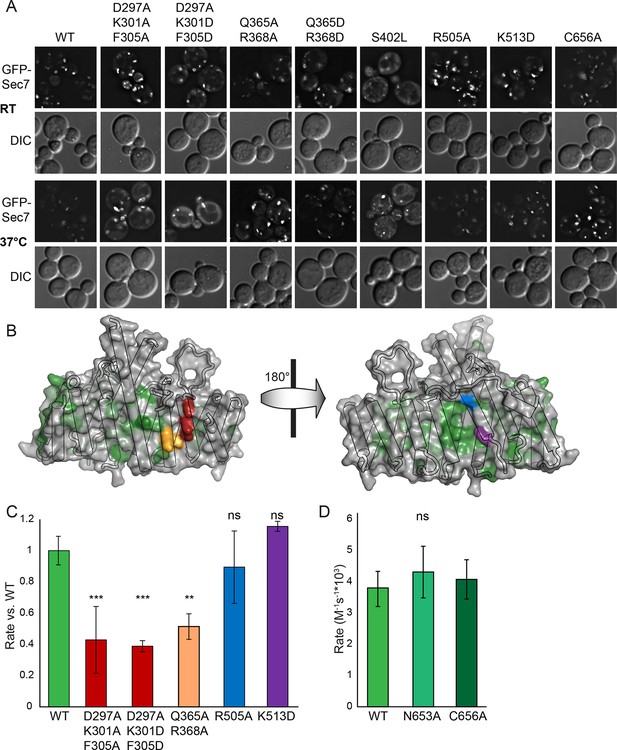
Helices 4 and 6 of the DCB/HUS domain mediate GEF stimulation.
(A) sec7Δ/arf1Δ strains bearing the indicated GFP-Sec7 alleles on a centromeric plasmid expressed from the SEC7 promoter were imaged at permissive and restrictive temperatures. (B) Surface residue conservation is shown on the basis of a 361-sequence MUSCLE alignment comprising all BLAST hits of the T. terrestris Sec7 N-terminus following removal of incomplete sequences and sequences with more than 95% pairwise identity. Green represents conservation. Residues mutated for biochemical assays are shown in colors matching the resultant bars. A closer view of the residue mutated in sec7-1 is shown in supplement 1. (C) Mutants purifiable as S. cerevisiae Sec7ΔC constructs were assayed for rate of nucleotide exchange of myristoylated Arf1 in the presence of liposomes. Activity of the same mutants toward ΔN17Arf1 in the absence of liposomes is shown in supplement 2. (D) The two HUS-box mutants purifiable as S. cerevisiae Sec7ΔC constructs were assayed for rate of nucleotide exchange of myristoylated Arf1 in the presence of liposomes. Viability and in vivo stability are assessed in supplements 3 and 4.
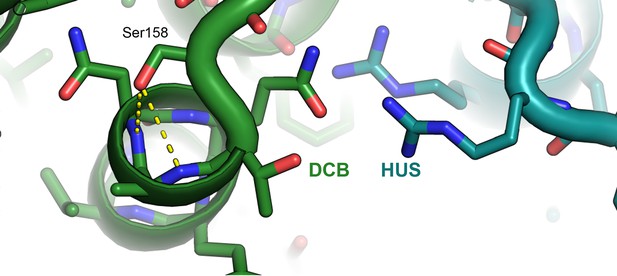
Atomic basis of the sec7-1 phenotype.
The temperature sensitive allele sec7-1 represents an S402L mutation, corresponding to T. terrestris Sec7 residue S158. This serine stabilizes a loop and N-terminal helical cap near the interface between the DCB and HUS regions.
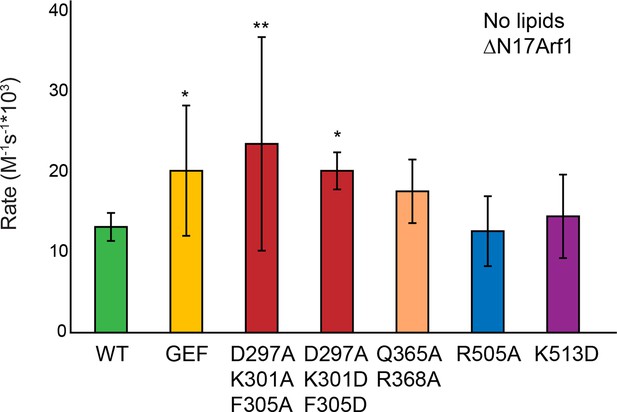
Sec7ΔC mutant activity in the absence of membranes.
Purifiable mutants in the S. cerevisiae Sec7ΔC construct were assayed for rate of nucleotide exchange of ΔN17Arf1 in the absence of membranes.
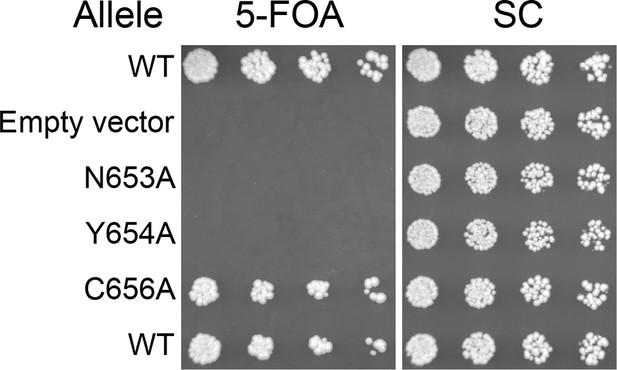
Viability of HUS box mutants.
Missense mutations in the HUS box were tested for viability at room temperature by plasmid shuffle, spotted in half-log dilutions left to right.
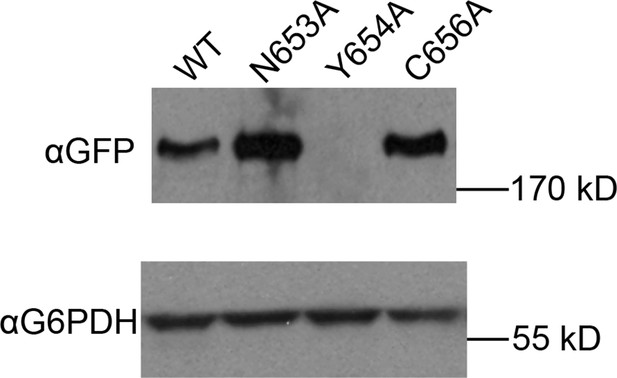
Expression of HUS box mutants.
Expression of the indicated GFP-Sec7f alleles was assayed by anti-GFP immunoblot.
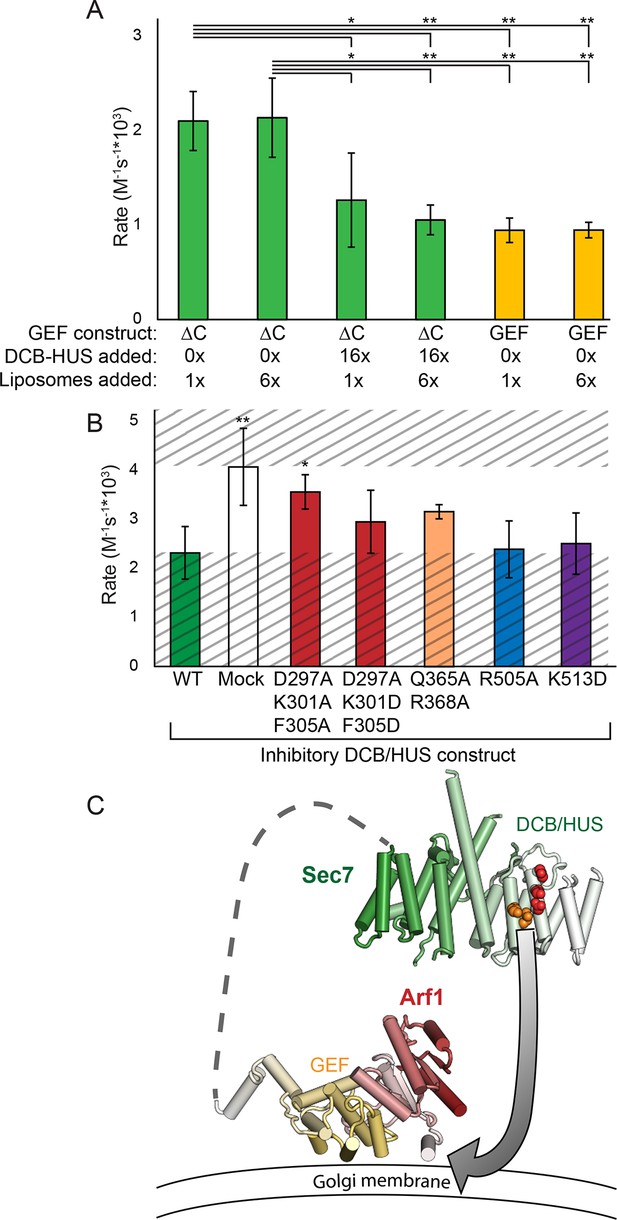
The DCB/HUS domain can inhibit GEF activity in trans.
(A) S. cerevisiae Sec7ΔC and isolated GEF constructs were assayed for rate of nucleotide exchange of myristoylated Arf1 in the presence of liposomes, with 16-fold excess DCB/HUS construct or sixfold additional liposomes added as indicated. (B) Wild-type S. cerevisiae Sec7ΔC was assayed for nucleotide exchange of myristoylated Arf1 in the presence of liposomes and a 12-fold excess of DCB/HUS constructs bearing the indicated mutations. The range of activity of interest is bounded by the WT and mock rates and is left unshaded. (C) A speculative model of DCB/HUS domain and GEF domain cooperation in Arf1 activation.
Tables
Data collection and refinement statistics
| T. terrestris Sec7 DCB/HUS domain (residues 1-458) | |
|---|---|
| Wavelength (Å) | 0.987 |
| Resolution range (Å) | 50 - 2.6 (2.64–2.6) |
| Space group | P 21 21 21 |
| Unit cell | a=62.472Å b=132.024Å c=247.664Å α=β=γ=90° |
| Total reflections | 569136 |
| Unique reflections | 59606 |
| Multiplicity | 9.5 (4.7) |
| Completeness (%) | 98.77 (88.46) |
| Mean I/sigma(I) | 8.46 (1.48) |
| Wilson B-factor | 65.26 |
| R-work | 0.2119 (0.3113) |
| R-free | 0.2568 (0.3665) |
| Number of atoms | 10944 |
| Macromolecules | 10887 |
| Water | 57 |
| Protein residues | 1383 |
| RMS(bonds) | 0.009 |
| RMS(angles) | 1.23 |
| Ramachandran favored (%) | 98 |
| Ramachandran outliers (%) | 0.075 |
| Clashscore | 7.63 |
| Average B-factor | 89.7 |
| Macromolecules | 89.8 |
| Solvent | 60.3 |
Additional files
-
Supplementary file 1
T. terrestris intron assignment
C. thermophilum, M. thermophila, and T. terrestris Sec7 genomic sequences, each containing a single annotated intron in the Sec7ΔC region, are aligned with annotated introns shown in lowercase. Conservation suggests that the T. terrestris intron should instead match that of the other two species; the intron assignment assumed for this work is highlighted in gray, and this correction to the construct was required for its expression (not shown).
- https://doi.org/10.7554/eLife.12411.027
-
Supplementary file 2
Plasmids and strain tables
- https://doi.org/10.7554/eLife.12411.028
-
Supplementary file 3
Estimation of exocytic Arf1 flux.
Values for Arf1 trafficking calculations are provided.
- https://doi.org/10.7554/eLife.12411.029





