Inhibiting poly(ADP-ribosylation) improves axon regeneration
Figures
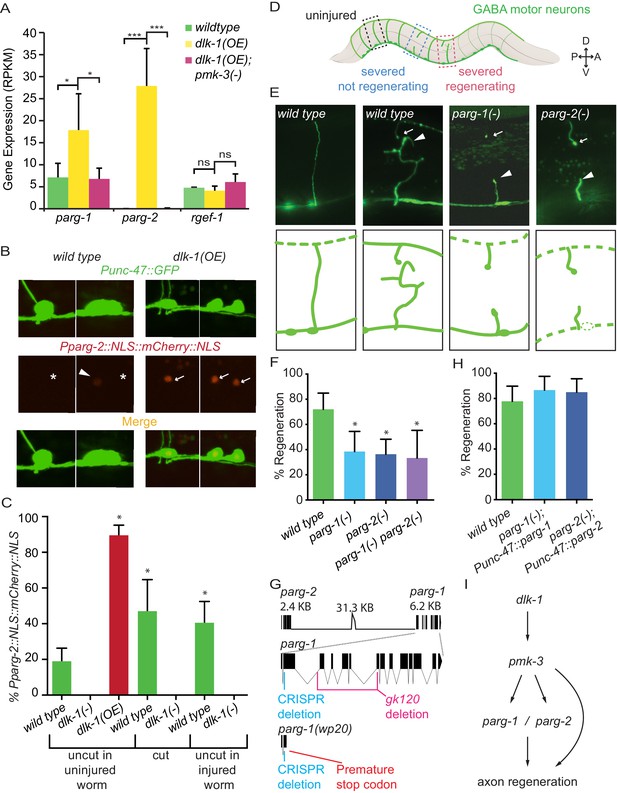
PARG genes regulate axon regeneration.
(A) dlk-1 overexpression upregulates parg-1 and parg-2 expression levels in GABA neurons. The upregulation is suppressed by loss of pmk-3 function. rgef-1 (a pan-neuronal Ras nucleotide exchange factor) expression levels are not affected by manipulations of the dlk-1 pathway (*p<0.05, ***p<0.01, two-way ANOVA, Bonferroni post-test). (B–C) dlk-1 regulates expression of nuclear-localized mCherry driven by the parg-2 promoter. Pparg-2::NLS::mCherry::NLS was observed (arrows) in 90% of nuclei of GABA neurons in dlk-1(OE) animals and in 19% of GABA neurons in wild type animals (asterisks). GABA neurons express GABA neuron-specific GFP marker, Punc-47::GFP. (C) parg-2 expression was significantly increased in both severed axons and neighboring uncut axons relative to axons in uninjured wild type animals. parg-2 was not expressed in dlk-1(lf) axons, whether severed or intact. (*p<0.05, Fisher’s exact test, relative to wild type, n = 111, 20, 115, 34, 18, 69, 36). (D) The GABA motor nervous system of C. elegans. GFP-labeled axons were severed with a pulsed laser at the midline (dark brown line) and scored for regeneration. (E) Representative micrographs of uninjured wild type, severed wild type, severed parg-1(-), and severed parg-2(-) GABA axons. Each carry the oxIs12 transgene which drives GFP expression in GABA neurons. Arrowheads and arrows indicate proximal and distal stumps, respectively. (F) Axon regeneration is significantly reduced in parg-1(-), parg-2(-) and parg-1(-) parg-2(-) mutants compared to wild type animals (*p<0.05, Fisher’s exact test, relative to wild type, n = 50, 39, 67, 21). (G) parg-1 and parg-2 are closely linked on chromosome IV, making construction of a double mutant difficult. To create a double parg-1 parg-2 mutant, parg-1 was mutated with CRISPR in a parg-2(lf) background. The resulting frameshift mutation (wp20) truncates PARP-1 earlier than the canonical gk120 deletion allele. (H) Expression of parg-1 or parg-2 in GABA motor neurons rescued axon regeneration in parg-1(lf) and parg-2(lf) mutants, respectively. (I) Model of PARG function in axon regeneration.
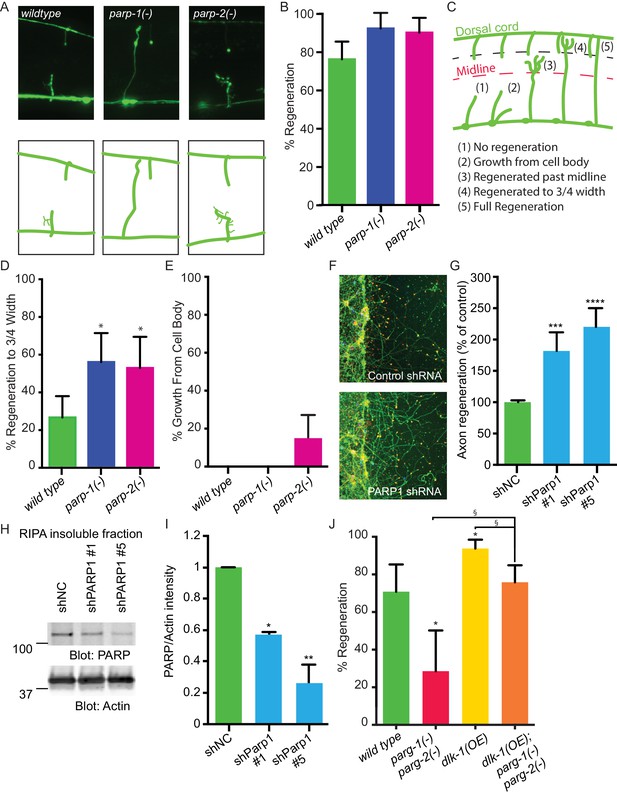
PARPs inhibit axon regeneration.
(A) Representative micrographs of severed wild type, parp-1(lf), and parp-2(lf) GABA motor neurons. (B) Axon regeneration in parp-1(lf) and parp-2(lf) mutants compared to wild type animals (*p<0.05, Fisher’s exact test, n = 84, 26, 61). (C) Cut axons (1) are scored for the distance they extend towards their targets in the dorsal nerve cord (2, 3, 4, 5). (D) Axon regeneration to at least 3/4 of the distance to the dorsal cord (4) is significantly increased in parp-1(lf) and parp-2(lf) mutants relative to wild type animals (*p<0.01, Fisher’s exact test, n = 61, 43, 38). (E) Axon regeneration from the cell body (2) is seen in parp-2(lf) mutants (n = 47, 32, 34). (F) Representative micrographs of injured cortical neurons exposed to negative control shRNA or PARP1 shRNA. (G) Axon regeneration is increased in murine cortical neurons lacking PARP1 (***p<0.001, ****p<0.0001, Anova with Tukey’s multiple comparisons test, n = 108, 8, 8). Axon regeneration was measured in injured cortical neurons exposed to non-coding negative control shRNA (shNC) or either of two unique PARP1 shRNAs. (H, I) Exposure to either shPARP significantly reduced PARP levels in cortical neurons relative to PARP levels in cortical neurons exposed to negative control (shNC) lentivirus (*p<0.05, **p<0.005, Anova with Tukey’s multiple comparisons test). (J) parg-1 and parg-2 loss of function incompletely suppress the increase in regeneration conferred by dlk-1(OE) (*p<0.05, relative to wild type, §p<0.05, relative to dlk-1(OE), Fisher’s exact test, n = 24, 21, 48, 62), indicating the PARGs regulate regeneration downstream of dlk-1 with at least one parallel pathway.
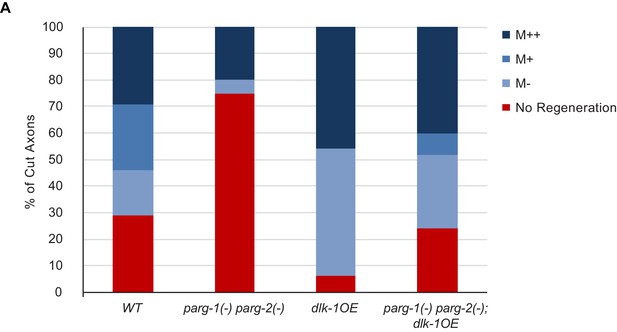
Detailed characterization of regeneration in dlk-1(OE) and parg-2(-) parg-2(-) mutants.
For each severed axon represented in Figure 2H, we determined whether a given axon regenerated below the midline of the worm (M-), beyond the midline (M+), at least 3/4 of the distance to the dorsal cord (M++). The greatest difference in regenerative ability between the genotypes is in overall regeneration, represented by combining the blue bars for each genotype.
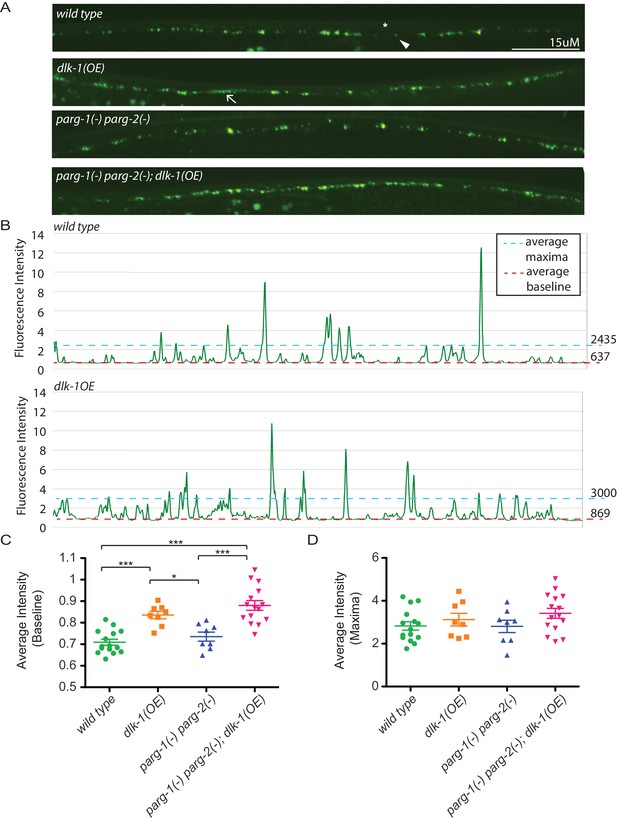
Loss of parg-1 and parg-2 function does not suppress mislocalizion of presynaptic active zones caused by dlk-1 overexpression.
(A) Dorsal nerve cords of wild type, dlk-1(OE), parg-1(lf) parg-2(lf), and dlk-1(OE); parg-1(lf) parg-2(lf) animals. All animals express the presynaptic active zone marker SYD-2::GFP in their GABA neurons. SYD-2::GFP is expressed in discrete puncta (arrowhead) in wild type animals and is not expressed continuously along the dorsal cord (asterisk). Conversely, SYD-2::GFP is expressed in a diffuse pattern (arrow) in dlk-1(OE) animals. (B) Average maxima and average baseline fluorescence are calculated along line scans of each dorsal cord and represented in (C). (C) dlk-1 overexpression disrupts SYD-2::GFP expression in GABA neurons and results in higher baseline fluorescence compared to wild type. Loss of parg function does not affect localization of SYD-2::GFP nor does it suppress the mislocalization caused by dlk-1(OE) (*p<0.05, ***p<0.01, multiple ANOVA, Bonferroni post-test). (D) There are no significant differences in average maxima between genotypes. Sample size is 15, 8, 8, and 15 animals for wild type, dlk-1(OE), parg-1(lf) parg-2(lf), and dlk-1(OE); parg-1(lf) parg-2(lf) animals, respectively.
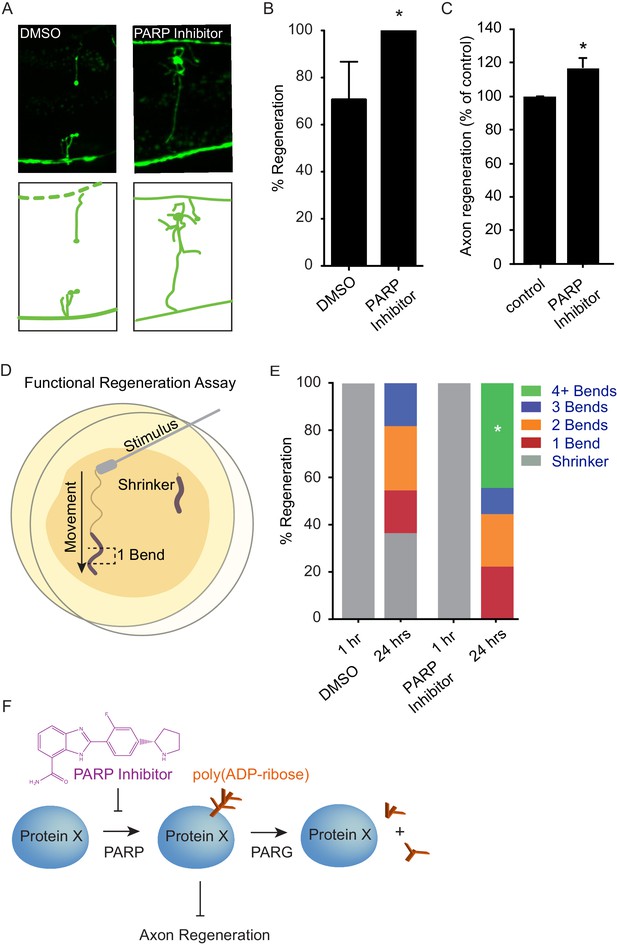
Chemical PARP inhibition enhances axon regeneration post-injury.
(A) Micrographs of regenerating axons placed on plates containing DMS0 or PARP inhibitor (A966492, Selleckchem, 100 µM) immediately after surgery. (B) Acute chemical inhibition of PARP function enhances regeneration (*p<0.05, Fisher’s exact test, n = 34 and 19 axons severed in animals exposed to DMSO or PARP inhibitor, respectively). (C) Axon regeneration is increased in murine cortical neurons exposed to chemical PARP inhibitor A966492 (*p=0.0149, Student’s t-test, n = 10). (D) To assess functional regeneration, all GABA neurons were severed and animals were assessed for their ability to reverse in response to a touch on the nose from a platinum wire. (E) One hour after all GABA neurons are severed, animals are incapable of reversing in response to a touch on the nose (shrinker). As functional connections are regenerated, animals recovered on PARP inhibitor displayed more backward movement than those recovered on DMSO (measured as number of body bends). Significantly more animals on PARP inhibitors recovered wild type function (4+ body bends, *p<0.05, Fisher’s exact test, n = 11 and 9 animals exposed to DMSO or PARP inhibitor, respectively). (F) The balance between PARP and PARG regulates axon regeneration and is altered by chemical PARP inhibitors.
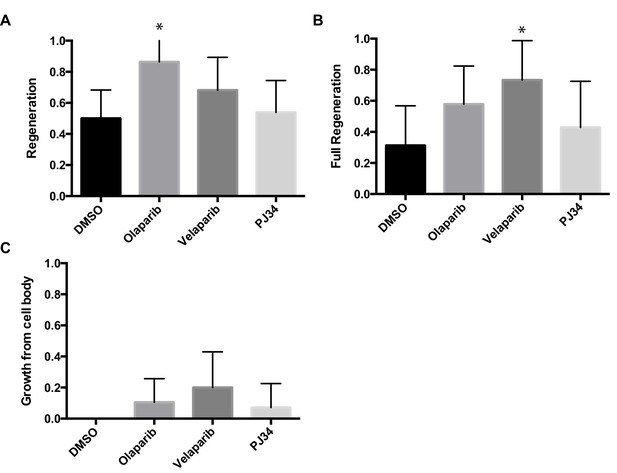
Chemical PARP inhibitors have different effects on axon regeneration.
Axon regeneration frequencies of wildtype animals placed on DMSO, Olaparib, Velaparib, or PJ34, post-axotomy. Regeneration was scored as (A) proportion of cut axons that regenerated, (B) proportion of regenerating axons that fully regenerated to the dorsal nerve cord (see Figure 2C), and (C) proportion of severed neurons that extended a secondary process rather than regenerate the severed axon.*p<0.05, Fisher’s exact test, relative to wild type; error bars represent 95% confidence intervals; n = 32, 22, 22, 26).





