Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection
Figures
Replication phenotypes of IAV WT and compositionally altered mutants.
(A) Replication kinetics of IAV in a multi-cycle replication assay; MDCK cells were infected at an MOI of 0.01 and supernatant assayed for IAV RNA at three post-infection time points; y-axis records infectivity of supernatant on titration in MDCK cells. Error bars show SEM of 3 replicate assays. A single replication cycle assay is shown in Figure 1—figure supplement 1. (B) Infectivity titres of supernatants collected at 24 hr. Bars show geometric titres of 5 replicate cultures of WT, CDLR, CpG-high and UpA-high variants; error bars show SEMs. The significance of titres differences was determined by Kruskall-Wallis non-parametric test; p values shown above bars with significant values highlighted in bold. (C) Mean plaque diameters of approximately 30 plaques of WT and mutant IAV variants; error bars show SEMs. (D, E) HA and RNA/infectivity ratios of WT and permuted, CpG-high and UpA-high mutants in MDCK cells; bar heights show mean values from 2–3 replicate assays; error bars show standard errors of the mean (SEM). (F) Infectivity titres of lung homogenates collected at days 3, 6 and 14 (experiment 2; Figure 2) and day 7 from mice infected with 20 PFU (experiment 3) from inoculated mice determined by titration on MDCK cells. Bar heights show mean values from cohorts of 4–6 mice; error bars show SEM. Synonymous site variability and composition analysis of IAV segment 5 is shown in Figure 1—figure supplement 1. Replication kinetics of IAV in a single cycle / high MOI replication assay is shown in Figure 1—figure supplement 2. Expression of IAV viral proteins M2, NS1, NA and PB2 relative to that of NP in different IAV mutants at 6 hr post-infection is shown in Figure 1—figure supplement 3. Detection of IAV viral proteins HA1, M1 and HA2 relative to that of NP in purified virions from different IAV mutants is shown in Figure 1—figure supplement 4. Ratio of segment 5 and segment 2 RNA sequences in purified virions is shown in Figure 1—figure supplement 5. Pairwise comparisons of the replication fitness of the mutants by competition assays is shown in Figure 1—figure supplement 6.
Variability and composition analysis of IAV segment 5.
(A) Variability at synonymous sites of selected subtypes of IAV (H1N1, H3N2, avian-derived variants H5, H6 and H9) in the coding region of segment 5 recorded as mean within-group pairwise p-distance. Sequences were scanned using window size of 30 codons, incrementing by 2 codons. (B) CpG and UpA dinucleotide frequencies in the coding region of segment 5, using mean values of 120 bases incrementing by 1 base per data point.
Single replication cycle kinetics of IAV infected an am MOI of 5 Replication kinetics of IAV in a single-cycle replication assay; MDCK cells were infected at an MOI of 5 and supernatant assayed for IAV RNA at 5 post-infection time points; y-axis records infectivity of supernatant on titration in MDCK cells.
https://doi.org/10.7554/eLife.12735.005
Expression of IAV viral proteins M2, NS1, NA and PB2 relative to that of NP in different IAV mutants.
Frequency of MDCK cells expressing different viral proteins 6 hr after infection with WT and mitant strains of IAV. Frequencies of infected cells were compared to those expressing the NP protein encoded by segment 5.

Detection of IAV viral proteins HA1, M1 and HA2 relative to that of NP in purified virions of different IAV mutants.
(A) PAGE of purified IAV virions derived from egg cultures of WT and mutant IAV strains. The major contaminant band in the CDLR preparation is most likely ovalbumin; its presence did not influence our ability to quantify IAV protein. (B) Viral proteins were quantified by densitometry and amounts relative to that of the NP protein (encoded by segment 5) plotted on y-axis.
Ratio of segment 5 and segment 2 RNA sequences in purified virions Quantitation of segment 5 and segment 2 RNA by qPCR in purified virions of WT and mutant IAV strains.
The y-axis records mean values of two biological replicates with values normalised to the ratio observed in WT virus; error bars show SEMs.
Pairwise comparisons of the replication fitness of the mutants by competition assays.
(A) Assay design and relative quantitation of virus pairs using restriction enzyme digestion of PCR product to differentiate amplicon sequences. (B) Summary results of pairwise comparisons. Cells were filled to illustrate diagrammatically the frequency of variants listed in columns; values and passage numbers indicated in boxes. The relative fitness ranking was PR8-WT ≥ CDLR >> UpA-high = CpG-high.
Infection outcomes and protective immunity in mice infected with IAV.
(A, B) Weights of mice (proportion of starting weight) of mice inoculated with 200 PFU of IAV WT and mutant strains with altered dinucleotide compositions. Deaths of individual mice are shown underneath the x-axis using the same colour coding. (C) Weight loss of mice infected with different inoculum doses (20–200 PFU) of the CpG-high IAV mutant (D) weight loss in the mice previously infected with CpG-high IAV (in Figure 2C) with a 200 PFU WT challenge dose at day 21 after the original inoculation. In all graphs, error bars show SEMs of 4–6 mice per group.
Cytopathology and innate immune responses to IAV infection in mice.
(A) Representative lung sections from mice infected with WT (a, b, e, f) and CpG-high (c, d, g, h) IAV variants at days 3 (a-d) and 14 (e-h) post infection. At day 3 prominent peribronchiolar and perivascular accumulation of inflammatory cells are present in WT infected mice (a), with moderate to marked, multifocal to coalescing airway epithelial cell necrosis (b). The inflammatory and necrotic lesions in this acute phase of disease are less severe in the CpG-high infected mice (c, d). At day 14 WT-infected mice showed marked peribronchial and perivascular lymphohistiocytic inflammation (e), epithelial regeneration and prominent type II pneumocyte proliferation (f). Again, the lesions in the CpG-high infected mice during this repair stage of disease were less severe (g, h). Bars in figures a, c, e, and g represent 200 µm. Bars in figure b, d, f, and h represent 20 µm. (B, C and D) Blinded histological scoring of (B) inflammation, (C) necrosis, and (D) repair processes in sections of lung from mice infected with different IAV strains. Bar heights show mean values from 4–6 mice per group scored from 0–3 for severity of (B) inflammation (perivascular, peribronchiolar and interstitial), (C) epithelial and interstitial necrosis, and (D) epithelial cell repair and type II pneumocyte proliferation. All average scores were normalised by the area of lung affected in the section. The significance of differences in pathology severity between viruses was determined by Kruskall-Wallis non-parametric test (combining WT and CDLR scores); p values shown above bars with significant values highlighted in bold. Error bars show SEM. Induction of individual cytokine in mice infected with IAV infection at different time point post-inoculation is shown in Figure 3—figure supplement 1 and 3. Induction of interferon-β mRNA in lung samples is shown in Figure 3—figure supplement 2.
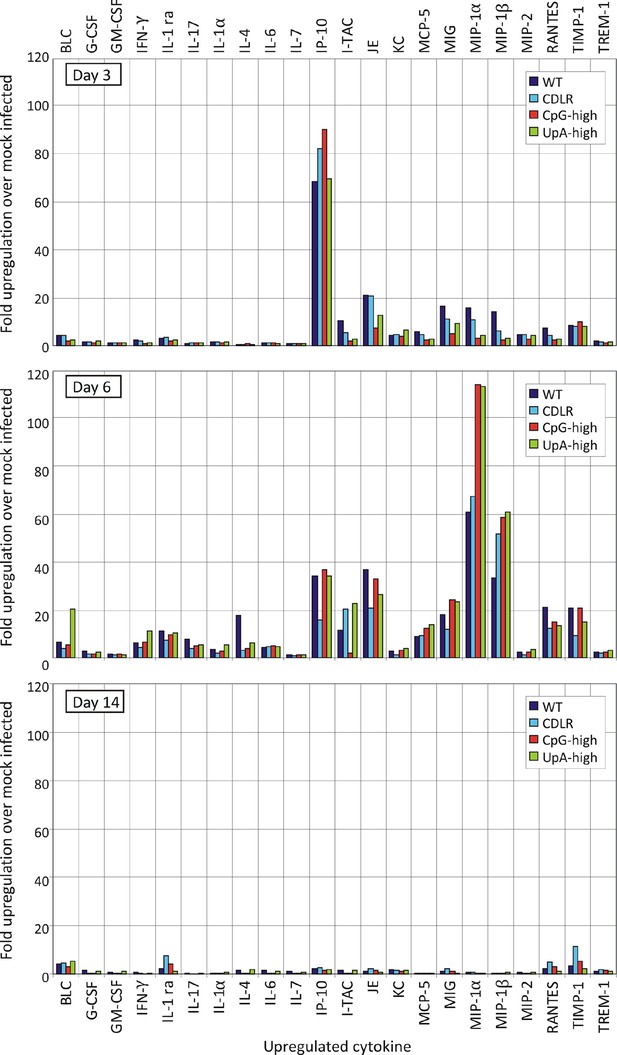
Cytokine response to IAV infection at different time point post-inoculation.
Induction of individual cytokines in pooled lung samples (n = 6) collected at days 3, 6 and 14 post-inoculation. The y-axis records fold-induction over cytokine level detected in uninfected mice; only cytokines showing significant (>two fold induction at any time point) are shown on the graph; no induction of the following cytokines were recorded: C5/C5a, Eotaxin, I-309, IL-10, IL-12 p70, IL-13, IL-16, IL-1α, IL-2, IL-23, IL-27, IL-3, IL-5, M-CSF, SDF-1, sICAM-1, TARC and TNF-α.
Induction of interferon-β mRNA in lung samples.
Quantitation of IFN-β mRNA in lungs of infected mice at days 3 and 6 by qPCR. Bars record mean levels; error bars show SEM. IFN-β was undetectable in all lung samples collected at day 14 (data not shown).
Cytokine responses in lungs of mice infected in experiment 1 (day 5).
Induction of individual cytokines using a standard mouse panel in pooled lung samples (n = 6) collected at days 3, 6 and 14 post-inoculation. The y-axis records fold-induction over cytokine level detected in uninfected mice; only cytokines showing significant (>two fold induction at any time point are shown on the graph; no induction of the following cytokines were recorded: C5/C5a, Eotaxin, I-309, IL-10, IL-12 p70, IL-13, IL-16, IL-1α, IL-2, IL-23, IL-27, IL-3, IL-5, M-CSF, SDF-1, sICAM-1, TARC and TNF-α.
Adaptive immune response (T cell and neutralising antibody) after infection with wild-type and mutant strains of IAV.
(A) Representative FACS plots showing the percentage of IFN-γ producing cells in mice infected with 200 PFU of IAV in the CD8+ T cell population after peptide stimulation. (B, C) Mean frequencies of CD8+ T lymphocytes expressing IFN- γ, TNF-α or IL-2 from spleen and lung at day 21 and neutralising antibody titres at days 6 and 21 post-infection with 200 PFU or 20 PFU of IAV respectively. Bars show mean frequencies of lymphocyte subsets and antibody titres from 4–6 mice per group; error bars show SEM. The gating strategy to identify CD8 lymphocytes is shown in Figure 4—figure supplement 1. Reactivity to individual peptides is shown in Figure 4—figure supplement 2.
Gating strategy to identify and quantify lymphocytes producing IFN-γ.
Gating strategy used to isolate the population of CD8 T cell IFN-γ producers after peptide stimulation.

ELISPOT analysis of T cell reactivity to individual IAV peptides.
The number of IFN-γ reactive SFU per 106 measured using ELISPOT following overnight stimulation of thawed splenocytes with peptides or media from mice immunised 21 days earlier with 200pfu of the indicated viruses. Bars show mean frequencies of lymphocyte subsets from 3–4 mice per group. Error bars show standard deviation.
Tables
Composition and coding parameters of the mutated region of segment 5.
| Subs.a | C+G% | CpG | ΔCpGb | CpG-O/Ec | UpA | ΔUpAb | UpA-O/E | CAI | CPSd | |
|---|---|---|---|---|---|---|---|---|---|---|
| PR8 WT | --- | 0.46 | 28 | --- | 0.43 | 43 | --- | 0.49 | 0.745 | 0.005 |
| CDLR | 134 | 0.46 | 28 | 0 | 0.43 | 43 | 0 | 0.49 | 0.745 | 0.011 |
| CpG-high | 233 | 0.46 | 114 | +86 | 1.63 | 45 | +2 | 0.51 | 0.611 | -0.011 |
| UpA-high | 199 | 0.46 | 29 | +1 | 0.56 | 116 | +73 | 1.31 | 0.627 | -0.118 |
-
a Number of sequence changes from WT sequence
-
b Change in the numbers of CpG and UpA dinucleotides
-
c Observed to expected frequencies of CpG and UpA dinucleotides
-
dCalculated as previously described (Buchan et al., 2006)
Primers used for qPCR and for amplification of segment 5 for competition assays.
| Gene/Region | Aim | Primer type | Sequence |
|---|---|---|---|
| Seg 5 | Reverse Transcription | Sense | ATCATGGCGTCTCAAGGCAC |
| Seg 5 | Sequencing | Sense | GAATGCCACTGAAATCAGAG |
| Antisense | CGTCCGAGAGCTCGAAGACT | ||
| Seg 5 | Competition Assay | Sense | CCAGAATGCCACTGAAATCA |
| Antisense | CCTTGCATYAGMGAGCACAT | ||
| Seg 5 | RT-qPCR | Sense | ATCATGGCGTCTCAAGGCAC |
| Antisense | CCGACGGATGCTCTGATTTC | ||
| GAPDH | RT-qPCR | Sense | CTACCCCCAATGTGTCCGTCG |
| Antisense | GATGCCTGCTTCACCACCTTC | ||
| IFN-β | RT-qPCR | Sense | CACAGCCCTCTCCATCAACT |
| Antisense | GCATCTTCTCCGTCATCTCC |
List of enzymes used in pairwise competition assays.
| Virus 1* | Virus 2* | Restriction Enzyme | Digestion Site |
|---|---|---|---|
| WT | CDLR | Hpy188III | In CDLR |
| WT | CpGH | Hpy188III | In CpGH |
| WT | UpAH | Hpy188III | In UpAH |
| CpGH | UpAH | AhdI | In CpGH |
| CpGH | CDLR | AhdI | In CpGH |
| UpAH | CDLR | BsaHI | In UpAH |
-
*Virus pairs to be differentiated
Additional files
-
Supplementary file 1
Sequences of the mutated regions in segment 5 of IAV (CDLR, CpG-high and UpA-high) are provided in the Supplementary file.
- https://doi.org/10.7554/eLife.12735.020





